+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-8651 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the human ether-a-go-go related K+ channel | |||||||||
 Map data Map data | hERG truncation construct hERGT | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | K+ channel / PAS / CNBHD / voltage sensor / selectivity filter / TRANSPORT PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationinward rectifier potassium channel complex / negative regulation of potassium ion export across plasma membrane / regulation of heart rate by hormone / Phase 3 - rapid repolarisation / membrane repolarization during action potential / negative regulation of potassium ion transmembrane transport / membrane repolarization during ventricular cardiac muscle cell action potential / membrane repolarization during cardiac muscle cell action potential / potassium ion export across plasma membrane / voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization ...inward rectifier potassium channel complex / negative regulation of potassium ion export across plasma membrane / regulation of heart rate by hormone / Phase 3 - rapid repolarisation / membrane repolarization during action potential / negative regulation of potassium ion transmembrane transport / membrane repolarization during ventricular cardiac muscle cell action potential / membrane repolarization during cardiac muscle cell action potential / potassium ion export across plasma membrane / voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization / regulation of membrane repolarization / membrane repolarization / positive regulation of potassium ion transmembrane transport / delayed rectifier potassium channel activity / Voltage gated Potassium channels / inward rectifier potassium channel activity / potassium ion homeostasis / ventricular cardiac muscle cell action potential / membrane depolarization during action potential / regulation of ventricular cardiac muscle cell membrane repolarization / regulation of potassium ion transmembrane transport / potassium ion import across plasma membrane / regulation of heart rate by cardiac conduction / voltage-gated potassium channel activity / voltage-gated potassium channel complex / cardiac muscle contraction / potassium ion transmembrane transport / regulation of membrane potential / potassium ion transport / cellular response to xenobiotic stimulus / scaffold protein binding / transcription cis-regulatory region binding / ubiquitin protein ligase binding / positive regulation of DNA-templated transcription / perinuclear region of cytoplasm / cell surface / protein homodimerization activity / identical protein binding / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.8 Å | |||||||||
 Authors Authors | Wang WW / MacKinnon R | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Cell / Year: 2017 Journal: Cell / Year: 2017Title: Cryo-EM Structure of the Open Human Ether-à-go-go-Related K Channel hERG. Authors: Weiwei Wang / Roderick MacKinnon /  Abstract: The human ether-à-go-go-related potassium channel (hERG, Kv11.1) is a voltage-dependent channel known for its role in repolarizing the cardiac action potential. hERG alteration by mutation or ...The human ether-à-go-go-related potassium channel (hERG, Kv11.1) is a voltage-dependent channel known for its role in repolarizing the cardiac action potential. hERG alteration by mutation or pharmacological inhibition produces Long QT syndrome and the lethal cardiac arrhythmia torsade de pointes. We have determined the molecular structure of hERG to 3.8 Å using cryo-electron microscopy. In this structure, the voltage sensors adopt a depolarized conformation, and the pore is open. The central cavity has an atypically small central volume surrounded by four deep hydrophobic pockets, which may explain hERG's unusual sensitivity to many drugs. A subtle structural feature of the hERG selectivity filter might correlate with its fast inactivation rate, which is key to hERG's role in cardiac action potential repolarization. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_8651.map.gz emd_8651.map.gz | 55.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-8651-v30.xml emd-8651-v30.xml emd-8651.xml emd-8651.xml | 16.8 KB 16.8 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_8651.png emd_8651.png | 193.1 KB | ||
| Filedesc metadata |  emd-8651.cif.gz emd-8651.cif.gz | 6.7 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-8651 http://ftp.pdbj.org/pub/emdb/structures/EMD-8651 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8651 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8651 | HTTPS FTP |
-Validation report
| Summary document |  emd_8651_validation.pdf.gz emd_8651_validation.pdf.gz | 542.4 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_8651_full_validation.pdf.gz emd_8651_full_validation.pdf.gz | 542 KB | Display | |
| Data in XML |  emd_8651_validation.xml.gz emd_8651_validation.xml.gz | 6.5 KB | Display | |
| Data in CIF |  emd_8651_validation.cif.gz emd_8651_validation.cif.gz | 7.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-8651 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-8651 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-8651 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-8651 | HTTPS FTP |
-Related structure data
| Related structure data |  5va2MC  8650C  8652C  5va1C  5va3C C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_8651.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_8651.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | hERG truncation construct hERGT | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X: 0.57305 Å / Y: 0.57305 Å / Z: 0.46914 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
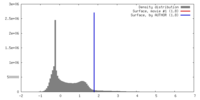
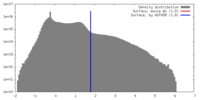
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : human ether-a-go-go related K+ channel hERG
| Entire | Name: human ether-a-go-go related K+ channel hERG |
|---|---|
| Components |
|
-Supramolecule #1: human ether-a-go-go related K+ channel hERG
| Supramolecule | Name: human ether-a-go-go related K+ channel hERG / type: organelle_or_cellular_component / ID: 1 / Parent: 0 / Macromolecule list: all Details: Truncated hERG construct hERGTs (amino acid residues 141-350 and 871-1005 deleted) |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Potassium voltage-gated channel subfamily H member 2
| Macromolecule | Name: Potassium voltage-gated channel subfamily H member 2 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 92.280539 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MPVRRGHVAP QNTFLDTIIR KFEGQSRKFI IANARVENCA VIYCNDGFCE LCGYSRAEVM QRPCTCDFLH GPRTQRRAAA QIAQALLGA EERKVEIAFY RKDGSCFLCL VDVVPVKNED GAVIMFILNF EVVMEKDMVG SSPTSDREII APKIKERTHN V TEKVTQVL ...String: MPVRRGHVAP QNTFLDTIIR KFEGQSRKFI IANARVENCA VIYCNDGFCE LCGYSRAEVM QRPCTCDFLH GPRTQRRAAA QIAQALLGA EERKVEIAFY RKDGSCFLCL VDVVPVKNED GAVIMFILNF EVVMEKDMVG SSPTSDREII APKIKERTHN V TEKVTQVL SLGADVLPEY KLQAPRIHRW TILHYSPFKA VWDWLILLLV IYTAVFTPYS AAFLLKETEE GPPATECGYA CQ PLAVVDL IVDIMFIVDI LINFRTTYVN ANEEVVSHPG RIAVHYFKGW FLIDMVAAIP FDLLIFGSGS EELIGLLKTA RLL RLVRVA RKLDRYSEYG AAVLFLLMCT FALIAHWLAC IWYAIGNMEQ PHMDSRIGWL HNLGDQIGKP YNSSGLGGPS IKDK YVTAL YFTFSSLTSV GFGNVSPNTN SEKIFSICVM LIGSLMYASI FGNVSAIIQR LYSGTARYHT QMLRVREFIR FHQIP NPLR QRLEEYFQHA WSYTNGIDMN AVLKGFPECL QADICLHLNR SLLQHCKPFR GATKGCLRAL AMKFKTTHAP PGDTLV HAG DLLTALYFIS RGSIEILRGD VVVAILGKND IFGEPLNLYA RPGKSNGDVR ALTYCDLHKI HRDDLLEVLD MYPEFSD HF WSSLEITFNL RDTNMIPGGR QYQELPRCPA PTPSLLNIPL SSPGRRPRGD VESRLDALQR QLNRLETRLS ADMATVLQ L LQRQMTLVPP AYSAVTTPGP GPTSTSPLLP VSPLPTLTLD SLSQVSQFMA CEELPPGAPE LPQEGPTRRL SLPGQLGAL TSQPLHRHGS DPGSEASNSL EVLFQ UniProtKB: Voltage-gated inwardly rectifying potassium channel KCNH2, Voltage-gated inwardly rectifying potassium channel KCNH2, Voltage-gated inwardly rectifying potassium channel KCNH2 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 6 mg/mL | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.4 Component:
Details: pH 7.4, adjusted with NaOH | ||||||||||||||||||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 400 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: PLASMA CLEANING / Pretreatment - Time: 12 sec. / Pretreatment - Atmosphere: AIR / Pretreatment - Pressure: 101.325 kPa | ||||||||||||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 98 % / Chamber temperature: 298 K / Instrument: FEI VITROBOT MARK IV / Details: one blot: 3 second blot time, 0 blot force. | ||||||||||||||||||||||||
| Details | A 1 mL peak fraction was collected and concentrated ~3x to obtain the final ~6 mg/mL sample. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Digitization - Dimensions - Width: 7420 pixel / Digitization - Dimensions - Height: 7676 pixel / Digitization - Frames/image: 1-50 / Number grids imaged: 1 / Number real images: 2443 / Average exposure time: 15.0 sec. / Average electron dose: 85.0 e/Å2 Details: 50 0.3-second frames were collected for each movie at a dose rate of ~1.8 e-/A2/frame |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 38461 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.5 µm / Nominal defocus min: 0.8 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: RECIPROCAL / Protocol: AB INITIO MODEL / Overall B value: 150 / Target criteria: Fourier Shell Correlation |
|---|---|
| Output model |  PDB-5va2: |
 Movie
Movie Controller
Controller











 Y (Sec.)
Y (Sec.) X (Row.)
X (Row.) Z (Col.)
Z (Col.)