[English] 日本語
 Yorodumi
Yorodumi- EMDB-8273: Human Islet Amyloid Polypeptide Segment 15-FLVHSSNNFGA-25 Determi... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-8273 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Human Islet Amyloid Polypeptide Segment 15-FLVHSSNNFGA-25 Determined by MicroED | |||||||||
 Map data Map data | Sigma-A-weighted 2Fo-Fc map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Amyloid / islet amyloid polypeptide / Type II Diabetes / Toxic Spine / MicroED / PROTEIN FIBRIL | |||||||||
| Function / homology |  Function and homology information Function and homology informationamylin receptor signaling pathway / Calcitonin-like ligand receptors / negative regulation of amyloid fibril formation / negative regulation of bone resorption / eating behavior / positive regulation of protein kinase A signaling / negative regulation of osteoclast differentiation / Regulation of gene expression in beta cells / negative regulation of protein-containing complex assembly / bone resorption ...amylin receptor signaling pathway / Calcitonin-like ligand receptors / negative regulation of amyloid fibril formation / negative regulation of bone resorption / eating behavior / positive regulation of protein kinase A signaling / negative regulation of osteoclast differentiation / Regulation of gene expression in beta cells / negative regulation of protein-containing complex assembly / bone resorption / sensory perception of pain / positive regulation of calcium-mediated signaling / osteoclast differentiation / hormone activity / cell-cell signaling / amyloid-beta binding / G alpha (s) signalling events / positive regulation of MAPK cascade / receptor ligand activity / positive regulation of apoptotic process / Amyloid fiber formation / signaling receptor binding / lipid binding / apoptotic process / signal transduction / extracellular space / extracellular region / identical protein binding Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | electron crystallography / cryo EM / Resolution: 1.4 Å | |||||||||
 Authors Authors | Krotee PAL / Rodriguez JA | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Elife / Year: 2017 Journal: Elife / Year: 2017Title: Atomic structures of fibrillar segments of hIAPP suggest tightly mated β-sheets are important for cytotoxicity. Authors: Pascal Krotee / Jose A Rodriguez / Michael R Sawaya / Duilio Cascio / Francis E Reyes / Dan Shi / Johan Hattne / Brent L Nannenga / Marie E Oskarsson / Stephan Philipp / Sarah Griner / Lin ...Authors: Pascal Krotee / Jose A Rodriguez / Michael R Sawaya / Duilio Cascio / Francis E Reyes / Dan Shi / Johan Hattne / Brent L Nannenga / Marie E Oskarsson / Stephan Philipp / Sarah Griner / Lin Jiang / Charles G Glabe / Gunilla T Westermark / Tamir Gonen / David S Eisenberg /    Abstract: hIAPP fibrils are associated with Type-II Diabetes, but the link of hIAPP structure to islet cell death remains elusive. Here we observe that hIAPP fibrils are cytotoxic to cultured pancreatic β- ...hIAPP fibrils are associated with Type-II Diabetes, but the link of hIAPP structure to islet cell death remains elusive. Here we observe that hIAPP fibrils are cytotoxic to cultured pancreatic β-cells, leading us to determine the structure and cytotoxicity of protein segments composing the amyloid spine of hIAPP. Using the cryoEM method MicroED, we discover that one segment, 19-29 S20G, forms pairs of β-sheets mated by a dry interface that share structural features with and are similarly cytotoxic to full-length hIAPP fibrils. In contrast, a second segment, 15-25 WT, forms non-toxic labile β-sheets. These segments possess different structures and cytotoxic effects, however, both can seed full-length hIAPP, and cause hIAPP to take on the cytotoxic and structural features of that segment. These results suggest that protein segment structures represent polymorphs of their parent protein and that segment 19-29 S20G may serve as a model for the toxic spine of hIAPP. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_8273.map.gz emd_8273.map.gz | 165.9 KB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-8273-v30.xml emd-8273-v30.xml emd-8273.xml emd-8273.xml | 15.1 KB 15.1 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_8273.png emd_8273.png | 116.4 KB | ||
| Filedesc metadata |  emd-8273.cif.gz emd-8273.cif.gz | 5.5 KB | ||
| Filedesc structureFactors |  emd_8273_sf.cif.gz emd_8273_sf.cif.gz | 39.4 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-8273 http://ftp.pdbj.org/pub/emdb/structures/EMD-8273 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8273 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8273 | HTTPS FTP |
-Validation report
| Summary document |  emd_8273_validation.pdf.gz emd_8273_validation.pdf.gz | 422.1 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_8273_full_validation.pdf.gz emd_8273_full_validation.pdf.gz | 421.7 KB | Display | |
| Data in XML |  emd_8273_validation.xml.gz emd_8273_validation.xml.gz | 4.1 KB | Display | |
| Data in CIF |  emd_8273_validation.cif.gz emd_8273_validation.cif.gz | 4.6 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-8273 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-8273 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-8273 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-8273 | HTTPS FTP |
-Related structure data
| Related structure data |  5ko0MC 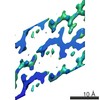 8272C  5knzC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_8273.map.gz / Format: CCP4 / Size: 179.7 KB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_8273.map.gz / Format: CCP4 / Size: 179.7 KB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sigma-A-weighted 2Fo-Fc map | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. generated in cubic-lattice coordinate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X: 0.449 Å / Y: 0.455 Å / Z: 0.453 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Amyloid fiber
| Entire | Name: Amyloid fiber |
|---|---|
| Components |
|
-Supramolecule #1: Amyloid fiber
| Supramolecule | Name: Amyloid fiber / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Molecular weight | Theoretical: 4.075 kDa/nm |
-Macromolecule #1: hIAPP(15-25)WT
| Macromolecule | Name: hIAPP(15-25)WT / type: protein_or_peptide / ID: 1 / Details: islet amyloid / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 1.193289 KDa |
| Sequence | String: FLVHSSNNFG A UniProtKB: Islet amyloid polypeptide |
-Macromolecule #2: THIOCYANATE ION
| Macromolecule | Name: THIOCYANATE ION / type: ligand / ID: 2 / Number of copies: 1 / Formula: SCN |
|---|---|
| Molecular weight | Theoretical: 58.082 Da |
| Chemical component information |  ChemComp-SCN: |
-Macromolecule #3: water
| Macromolecule | Name: water / type: ligand / ID: 3 / Number of copies: 6 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | electron crystallography |
| Aggregation state | 3D array |
- Sample preparation
Sample preparation
| Concentration | 20 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7 Component:
| |||||||||
| Grid | Model: Quantifoil R2/2 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY ARRAY / Support film - Film thickness: 30 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 20 sec. / Pretreatment - Atmosphere: AIR | |||||||||
| Vitrification | Cryogen name: ETHANE | |||||||||
| Crystal formation | Instrument: Hanging Drop Vapor Diffusion Tray / Atmosphere: Air, sealed chamber / Temperature: 277.0 K / Time: 7.0 DAY Details: 20 mg/mL Lyophilized peptide in ice-cold water. Crystals were grown using the hanging drop vapor diffusion method at 4 degrees C in 0.35 M sodium thiocyanate and 35% MPD. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI 20 |
|---|---|
| Temperature | Min: 100.0 K / Max: 100.0 K |
| Image recording | Film or detector model: TVIPS TEMCAM-F416 (4k x 4k) / Digitization - Dimensions - Width: 4096 pixel / Digitization - Dimensions - Height: 4096 pixel / Number grids imaged: 2 / Number diffraction images: 879 / Average exposure time: 2.0 sec. / Average electron dose: 0.01 e/Å2 Details: The detector was operated in rolling shutter mode with 2x2 pixel binning. |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: DIFFRACTION / Camera length: 1840 mm |
| Sample stage | Specimen holder model: GATAN 626 SINGLE TILT LIQUID NITROGEN CRYO TRANSFER HOLDER Cooling holder cryogen: NITROGEN |
- Image processing
Image processing
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 1.4 Å / Resolution method: DIFFRACTION PATTERN/LAYERLINES |
|---|---|
| Startup model | Type of model: PDB ENTRY / Details: 5.0E+61 |
| Molecular replacement | Software - Name: Phaser (ver. 2.5.6) |
| Crystallography statistics | Number intensities measured: 9014 / Number structure factors: 2180 / Fourier space coverage: 75 / R sym: 0.193 / R merge: 0.193 / Overall phase error: 0.01 / Overall phase residual: 0.01 / Phase error rejection criteria: 0 / High resolution: 1.4 Å Details: Phasing statistics are not applicable. No imaging was used. The phases were obtained using molecular replacement. Shell - Shell ID: 1 / Shell - High resolution: 1.4 Å / Shell - Low resolution: 22.0 Å / Shell - Number structure factors: 2180 / Shell - Phase residual: 0.01 / Shell - Fourier space coverage: 75 / Shell - Multiplicity: 4.1 |
-Atomic model buiding 1
| Refinement | Space: RECIPROCAL / Protocol: OTHER / Overall B value: 9.03 / Target criteria: maximum likelihood |
|---|---|
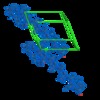
| Output model |  PDB-5ko0: |
 Movie
Movie Controller
Controller






 Y (Sec.)
Y (Sec.) X (Row.)
X (Row.) Z (Col.)
Z (Col.)