[English] 日本語
 Yorodumi
Yorodumi- EMDB-8098: Cryo-EM structure of GluN1/GluN2B NMDA receptor in the glutamate/... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-8098 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of GluN1/GluN2B NMDA receptor in the glutamate/glycine/Ro25-6981-bound conformation | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | ligand-gated ion channel / synaptic transmission / SIGNALING PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationNMDA glutamate receptor activity / NMDA selective glutamate receptor complex / ligand-gated sodium channel activity / response to zinc ion / response to magnesium ion / glutamate-gated calcium ion channel activity / ionotropic glutamate receptor signaling pathway / sodium ion transmembrane transport / excitatory postsynaptic potential / synaptic transmission, glutamatergic ...NMDA glutamate receptor activity / NMDA selective glutamate receptor complex / ligand-gated sodium channel activity / response to zinc ion / response to magnesium ion / glutamate-gated calcium ion channel activity / ionotropic glutamate receptor signaling pathway / sodium ion transmembrane transport / excitatory postsynaptic potential / synaptic transmission, glutamatergic / regulation of membrane potential / transmitter-gated monoatomic ion channel activity involved in regulation of postsynaptic membrane potential / regulation of synaptic plasticity / postsynaptic density membrane / calcium ion transmembrane transport / long-term synaptic potentiation / late endosome / signaling receptor activity / chemical synaptic transmission / postsynaptic membrane / lysosome / neuron projection / synapse / metal ion binding / plasma membrane Similarity search - Function | |||||||||
| Biological species | ||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 7.5 Å | |||||||||
 Authors Authors | Zhu S / Stein AR | |||||||||
 Citation Citation |  Journal: Cell / Year: 2016 Journal: Cell / Year: 2016Title: Mechanism of NMDA Receptor Inhibition and Activation. Authors: Shujia Zhu / Richard A Stein / Craig Yoshioka / Chia-Hsueh Lee / April Goehring / Hassane S Mchaourab / Eric Gouaux /  Abstract: N-methyl-D-aspartate receptors (NMDARs) are glutamate-gated, calcium-permeable ion channels that mediate synaptic transmission and underpin learning and memory. NMDAR dysfunction is directly ...N-methyl-D-aspartate receptors (NMDARs) are glutamate-gated, calcium-permeable ion channels that mediate synaptic transmission and underpin learning and memory. NMDAR dysfunction is directly implicated in diseases ranging from seizure to ischemia. Despite its fundamental importance, little is known about how the NMDAR transitions between inactive and active states and how small molecules inhibit or activate ion channel gating. Here, we report electron cryo-microscopy structures of the GluN1-GluN2B NMDA receptor in an ensemble of competitive antagonist-bound states, an agonist-bound form, and a state bound with agonists and the allosteric inhibitor Ro25-6981. Together with double electron-electron resonance experiments, we show how competitive antagonists rupture the ligand binding domain (LBD) gating "ring," how agonists retain the ring in a dimer-of-dimers configuration, and how allosteric inhibitors, acting within the amino terminal domain, further stabilize the LBD layer. These studies illuminate how the LBD gating ring is fundamental to signal transduction and gating in NMDARs. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_8098.map.gz emd_8098.map.gz | 29.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-8098-v30.xml emd-8098-v30.xml emd-8098.xml emd-8098.xml | 15.1 KB 15.1 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_8098.png emd_8098.png | 138.6 KB | ||
| Filedesc metadata |  emd-8098.cif.gz emd-8098.cif.gz | 6.7 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-8098 http://ftp.pdbj.org/pub/emdb/structures/EMD-8098 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8098 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8098 | HTTPS FTP |
-Related structure data
| Related structure data |  5iovMC  8097C  8101C  8102C  8103C  8104C  8105C  8106C  5iouC  5ipqC  5iprC  5ipsC  5iptC  5ipuC  5ipvC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_8098.map.gz / Format: CCP4 / Size: 32.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_8098.map.gz / Format: CCP4 / Size: 32.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
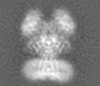
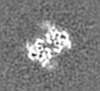
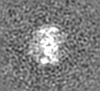
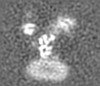
| Projections & slices | Image control
Images are generated by Spider. generated in cubic-lattice coordinate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.07 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : GluN1-GluN2B receptor in the complex with glycine and glutamate p...
| Entire | Name: GluN1-GluN2B receptor in the complex with glycine and glutamate plus allosteric inhibitor Ro25-6981 |
|---|---|
| Components |
|
-Supramolecule #1: GluN1-GluN2B receptor in the complex with glycine and glutamate p...
| Supramolecule | Name: GluN1-GluN2B receptor in the complex with glycine and glutamate plus allosteric inhibitor Ro25-6981 type: organelle_or_cellular_component / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism: |
-Macromolecule #1: N-methyl-D-aspartate receptor subunit NR1-8a
| Macromolecule | Name: N-methyl-D-aspartate receptor subunit NR1-8a / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: |
| Molecular weight | Theoretical: 92.651234 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: DPKIVNIGAV LSTKKHEQIF REAVNQANFF HFTRKIQLNA TSVTHRPNAI QMALSVCEDL ISSQVYAILV SHPPAPTDHL TPTPISYTA GFYRIPVIGL TTRMSIYSDK SIHLSFLRTV PPYSHQALVW FEMMRLFNWN HVILIVSDDH EGRAAQKKLE T LLEEKESK ...String: DPKIVNIGAV LSTKKHEQIF REAVNQANFF HFTRKIQLNA TSVTHRPNAI QMALSVCEDL ISSQVYAILV SHPPAPTDHL TPTPISYTA GFYRIPVIGL TTRMSIYSDK SIHLSFLRTV PPYSHQALVW FEMMRLFNWN HVILIVSDDH EGRAAQKKLE T LLEEKESK ADKVLQFEPG TKNLTALLLE AKELEARVII LSASEDDATA VYKSAAMLDM TGAGYVWLVG EREISGSALR YA PDGIIGL QLINGKNESA HISDAVAVVA QAIHELFEME QITDPPRGCV GNTNIWKTGP LFKRVLMSSK YPDGVTGRIE FNE DGDRKF AQYSIMNLQN RKLVQVGIFD GSYIIQNDRK IIWPGGETER PQGYQMSTRL KIVTIHQEPF VYVRPTTSDG TCRE EYTIN GDPIKKVICN GPDETIPGRP TVPQCCYGFC VDLLIKLARE MDFTYEVHLV ADGKFGTQER VNNSNAAAWN GMMGE LLSG QADMIVAPLT INNERAQYIE FSKPFKYQGL TILVKKEIPR STLDSFMQPF QSTLWLLVGL SVHVVAVMLY LLDRFS PFG RFEDALTLSS AMWFSWRVLL NSGLGEGAPR SFSARILGMV WAGFAMIIVA SYTANLAAFL VLRRPEERIT GINDPRL RN PSDKFIYATV KQSSVDIYFR RQVELSTMYR HMEKHNYESA AEAIQAVRDN KLHAFIWDSA VLEFEASQKC DLVTTGEL F FRSGFGIGMR KDSPWKQEVS LNILKSHENG FMEELDKTWV RYQECDSRSN APATLTFENM AGVFMLVAGG IVAGIFLIF IEIAYKSRAE AKRMKGLEVL FQ UniProtKB: Glutamate receptor ionotropic, NMDA 1 |
-Macromolecule #2: Ionotropic glutamate receptor subunit NR2B
| Macromolecule | Name: Ionotropic glutamate receptor subunit NR2B / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: |
| Molecular weight | Theoretical: 93.234742 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MRPTEACCYL KISLIILFYS RAYAQKHPNM DIAVILVGTT EEVAIKDVHE KDDFHHLPVT PRVELVTMQE SDPKSIITRI CDLMSDKKV QGVVFGDDTD QEAIAQILDF ISVQTLTPIL GIHGGSSMIM ADKEEASMFF QFGPSIEQQA SVMLNIMEEY D WYIFSIVT ...String: MRPTEACCYL KISLIILFYS RAYAQKHPNM DIAVILVGTT EEVAIKDVHE KDDFHHLPVT PRVELVTMQE SDPKSIITRI CDLMSDKKV QGVVFGDDTD QEAIAQILDF ISVQTLTPIL GIHGGSSMIM ADKEEASMFF QFGPSIEQQA SVMLNIMEEY D WYIFSIVT TYFPGYQDFE NKVRSTIENS FVGWELEEVI HLDMSLDDID SKIQNQLKKL QSPVILLYCT KEEATYIFEV AH SVGLTGY GFTWIVPSLV AGDTDTVPDE FPTGLISVSY DEWDYDLPAR VRDGIAIITT AASTMLSEHN SIPQSKSSCN NIQ ESRVYE AHMLKRYLIN VTFEGRDLSF SEDGYQMHPK LVIILLNQER KWERVGKYKD RSLKMWPVFD LYPNSEEHKD EHLS IVTLE EAPFVIVEDV DPLSGTCMRN TVPCRKQIRP ENRTEEGGNY IKRCCKGFCI DILKKIAKTV KFTYDLYLVT NGKHG KKIN GVWNGMIGEV VTKRAYMAVG SLTINEERSE VVDFSVPFIE TGISVMVSRS NGTVSPSAFL EPFSADVWVM MFVMLL IVS AVAVFVFEYF SPVGYNGPSF TIGKAIWLLW GLVFNNSLPV QNPKGTTSKI MVSVWAFFAV IFLASYTANL AAFMIQR RY VDQVSGLSDK KFQRPNDFSP AFRFGTVPNG STERNIRNNY LEMHSYMVKF NQRSVQDALL SLKSGKLDAF IYDAAVLN Y MAGRDEGCKL VTIGSGKVFA TTGYGIAIQK DSGWKRQVDL AILQLFGDGE MEELEALWLT GICHNEKNEV MSSQLDIDN MAGVFYMLAA AMALSLITFI MEHLF UniProtKB: Glutamate receptor ionotropic, NMDA 2B |
-Macromolecule #3: GLYCINE
| Macromolecule | Name: GLYCINE / type: ligand / ID: 3 / Number of copies: 2 / Formula: GLY |
|---|---|
| Molecular weight | Theoretical: 75.067 Da |
| Chemical component information |  ChemComp-GLY: |
-Macromolecule #4: 4-[(1R,2S)-3-(4-benzylpiperidin-1-yl)-1-hydroxy-2-methylpropyl]phenol
| Macromolecule | Name: 4-[(1R,2S)-3-(4-benzylpiperidin-1-yl)-1-hydroxy-2-methylpropyl]phenol type: ligand / ID: 4 / Number of copies: 2 / Formula: QEM |
|---|---|
| Molecular weight | Theoretical: 339.471 Da |
| Chemical component information | 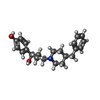 ChemComp-QEM: |
-Macromolecule #5: GLUTAMIC ACID
| Macromolecule | Name: GLUTAMIC ACID / type: ligand / ID: 5 / Number of copies: 2 / Formula: GLU |
|---|---|
| Molecular weight | Theoretical: 147.129 Da |
| Chemical component information |  ChemComp-GLU: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 4 mg/mL |
|---|---|
| Buffer | pH: 6.5 |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 18 K / Instrument: FEI VITROBOT MARK III |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Average electron dose: 10.2 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 0.01 mm / Nominal defocus max: 3.5 µm / Nominal defocus min: 1.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)