+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 7q1z | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
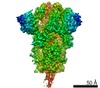
| Title | Structure of formaldehyde cross-linked SARS-CoV-2 S glycoprotein | ||||||||||||||||||||||||||||||||||||
 Components Components | Spike glycoprotein | ||||||||||||||||||||||||||||||||||||
 Keywords Keywords | VIRAL PROTEIN / SARS-CoV-2 / glycoprotein / immunization | ||||||||||||||||||||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationMaturation of spike protein / Translation of Structural Proteins / Virion Assembly and Release / host cell surface / host extracellular space / viral translation / symbiont-mediated-mediated suppression of host tetherin activity / Induction of Cell-Cell Fusion / structural constituent of virion / membrane fusion ...Maturation of spike protein / Translation of Structural Proteins / Virion Assembly and Release / host cell surface / host extracellular space / viral translation / symbiont-mediated-mediated suppression of host tetherin activity / Induction of Cell-Cell Fusion / structural constituent of virion / membrane fusion / entry receptor-mediated virion attachment to host cell / Attachment and Entry / host cell endoplasmic reticulum-Golgi intermediate compartment membrane / positive regulation of viral entry into host cell / receptor-mediated virion attachment to host cell / host cell surface receptor binding / symbiont-mediated suppression of host innate immune response / receptor ligand activity / endocytosis involved in viral entry into host cell / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / virion attachment to host cell / SARS-CoV-2 activates/modulates innate and adaptive immune responses / host cell plasma membrane / virion membrane / identical protein binding / membrane / plasma membrane Similarity search - Function | ||||||||||||||||||||||||||||||||||||
| Biological species |  | ||||||||||||||||||||||||||||||||||||
| Method | ELECTRON MICROSCOPY / single particle reconstruction / cryo EM / Resolution: 3.4 Å | ||||||||||||||||||||||||||||||||||||
 Authors Authors | Sulbaran, G. / Effantin, G. / Schoehn, G. / Weissenhorn, W. | ||||||||||||||||||||||||||||||||||||
| Funding support |  France, 2items France, 2items
| ||||||||||||||||||||||||||||||||||||
 Citation Citation |  Journal: Cell Rep Med / Year: 2022 Journal: Cell Rep Med / Year: 2022Title: Immunization with synthetic SARS-CoV-2 S glycoprotein virus-like particles protects macaques from infection. Authors: Guidenn Sulbaran / Pauline Maisonnasse / Axelle Amen / Gregory Effantin / Delphine Guilligay / Nathalie Dereuddre-Bosquet / Judith A Burger / Meliawati Poniman / Marloes Grobben / Marlyse ...Authors: Guidenn Sulbaran / Pauline Maisonnasse / Axelle Amen / Gregory Effantin / Delphine Guilligay / Nathalie Dereuddre-Bosquet / Judith A Burger / Meliawati Poniman / Marloes Grobben / Marlyse Buisson / Sebastian Dergan Dylon / Thibaut Naninck / Julien Lemaître / Wesley Gros / Anne-Sophie Gallouët / Romain Marlin / Camille Bouillier / Vanessa Contreras / Francis Relouzat / Daphna Fenel / Michel Thepaut / Isabelle Bally / Nicole Thielens / Franck Fieschi / Guy Schoehn / Sylvie van der Werf / Marit J van Gils / Rogier W Sanders / Pascal Poignard / Roger Le Grand / Winfried Weissenhorn /    Abstract: The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) pandemic has caused an ongoing global health crisis. Here, we present as a vaccine candidate synthetic SARS-CoV-2 spike (S) ...The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) pandemic has caused an ongoing global health crisis. Here, we present as a vaccine candidate synthetic SARS-CoV-2 spike (S) glycoprotein-coated lipid vesicles that resemble virus-like particles. Soluble S glycoprotein trimer stabilization by formaldehyde cross-linking introduces two major inter-protomer cross-links that keep all receptor-binding domains in the "down" conformation. Immunization of cynomolgus macaques with S coated onto lipid vesicles (S-LVs) induces high antibody titers with potent neutralizing activity against the vaccine strain, Alpha, Beta, and Gamma variants as well as T helper (Th)1 CD4-biased T cell responses. Although anti-receptor-binding domain (RBD)-specific antibody responses are initially predominant, the third immunization boosts significant non-RBD antibody titers. Challenging vaccinated animals with SARS-CoV-2 shows a complete protection through sterilizing immunity, which correlates with the presence of nasopharyngeal anti-S immunoglobulin G (IgG) and IgA titers. Thus, the S-LV approach is an efficient and safe vaccine candidate based on a proven classical approach for further development and clinical testing. #1:  Journal: Cell Rep Med / Year: 2022 Journal: Cell Rep Med / Year: 2022Title: Immunization with synthetic SARS-CoV-2 S glycoprotein virus-like particles protects macaques from infection. Authors: Guidenn Sulbaran / Pauline Maisonnasse / Axelle Amen / Gregory Effantin / Delphine Guilligay / Nathalie Dereuddre-Bosquet / Judith A Burger / Meliawati Poniman / Marloes Grobben / Marlyse ...Authors: Guidenn Sulbaran / Pauline Maisonnasse / Axelle Amen / Gregory Effantin / Delphine Guilligay / Nathalie Dereuddre-Bosquet / Judith A Burger / Meliawati Poniman / Marloes Grobben / Marlyse Buisson / Sebastian Dergan Dylon / Thibaut Naninck / Julien Lemaître / Wesley Gros / Anne-Sophie Gallouët / Romain Marlin / Camille Bouillier / Vanessa Contreras / Francis Relouzat / Daphna Fenel / Michel Thepaut / Isabelle Bally / Nicole Thielens / Franck Fieschi / Guy Schoehn / Sylvie van der Werf / Marit J van Gils / Rogier W Sanders / Pascal Poignard / Roger Le Grand / Winfried Weissenhorn /    Abstract: The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) pandemic has caused an ongoing global health crisis. Here, we present as a vaccine candidate synthetic SARS-CoV-2 spike (S) ...The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) pandemic has caused an ongoing global health crisis. Here, we present as a vaccine candidate synthetic SARS-CoV-2 spike (S) glycoprotein-coated lipid vesicles that resemble virus-like particles. Soluble S glycoprotein trimer stabilization by formaldehyde cross-linking introduces two major inter-protomer cross-links that keep all receptor-binding domains in the "down" conformation. Immunization of cynomolgus macaques with S coated onto lipid vesicles (S-LVs) induces high antibody titers with potent neutralizing activity against the vaccine strain, Alpha, Beta, and Gamma variants as well as T helper (Th)1 CD4-biased T cell responses. Although anti-receptor-binding domain (RBD)-specific antibody responses are initially predominant, the third immunization boosts significant non-RBD antibody titers. Challenging vaccinated animals with SARS-CoV-2 shows a complete protection through sterilizing immunity, which correlates with the presence of nasopharyngeal anti-S immunoglobulin G (IgG) and IgA titers. Thus, the S-LV approach is an efficient and safe vaccine candidate based on a proven classical approach for further development and clinical testing. #2:  Journal: Cell Rep Med / Year: 2022 Journal: Cell Rep Med / Year: 2022Title: Immunization with synthetic SARS-CoV-2 S glycoprotein virus-like particles protects macaques from infection. Authors: Sulbaran, G. / Effantin, G. / Schoehn, G. / Weissenhorn, W. | ||||||||||||||||||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  7q1z.cif.gz 7q1z.cif.gz | 540.6 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb7q1z.ent.gz pdb7q1z.ent.gz | 437.4 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  7q1z.json.gz 7q1z.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  7q1z_validation.pdf.gz 7q1z_validation.pdf.gz | 2.5 MB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  7q1z_full_validation.pdf.gz 7q1z_full_validation.pdf.gz | 2.5 MB | Display | |
| Data in XML |  7q1z_validation.xml.gz 7q1z_validation.xml.gz | 85.8 KB | Display | |
| Data in CIF |  7q1z_validation.cif.gz 7q1z_validation.cif.gz | 130.5 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/q1/7q1z https://data.pdbj.org/pub/pdb/validation_reports/q1/7q1z ftp://data.pdbj.org/pub/pdb/validation_reports/q1/7q1z ftp://data.pdbj.org/pub/pdb/validation_reports/q1/7q1z | HTTPS FTP |
-Related structure data
| Related structure data |  13776MC M: map data used to model this data C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
|
|---|---|
| 1 |
|
- Components
Components
| #1: Protein | Mass: 142399.375 Da / Num. of mol.: 3 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Gene: S, 2 / Production host:  Homo sapiens (human) / References: UniProt: P0DTC2 Homo sapiens (human) / References: UniProt: P0DTC2#2: Polysaccharide | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose Source method: isolated from a genetically manipulated source #3: Sugar | ChemComp-NAG / Has ligand of interest | Y | Has protein modification | Y | |
|---|
-Experimental details
-Experiment
| Experiment | Method: ELECTRON MICROSCOPY |
|---|---|
| EM experiment | Aggregation state: PARTICLE / 3D reconstruction method: single particle reconstruction |
- Sample preparation
Sample preparation
| Component | Name: Trimer of the SARS_CoV-2 S glycoprotein / Type: COMPLEX / Entity ID: #1 / Source: RECOMBINANT |
|---|---|
| Molecular weight | Experimental value: NO |
| Source (natural) | Organism:  |
| Source (recombinant) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Buffer solution | pH: 7.4 |
| Specimen | Embedding applied: NO / Shadowing applied: NO / Staining applied: NO / Vitrification applied: YES |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy imaging
Electron microscopy imaging
| Microscopy | Model: TFS GLACIOS |
|---|---|
| Electron gun | Electron source:  FIELD EMISSION GUN / Accelerating voltage: 200 kV / Illumination mode: FLOOD BEAM FIELD EMISSION GUN / Accelerating voltage: 200 kV / Illumination mode: FLOOD BEAM |
| Electron lens | Mode: BRIGHT FIELD |
| Image recording | Electron dose: 40 e/Å2 / Detector mode: COUNTING / Film or detector model: GATAN K2 SUMMIT (4k x 4k) |
- Processing
Processing
| Software | Name: PHENIX / Version: 1.19.2_4158: / Classification: refinement |
|---|---|
| EM software | Name: PHENIX / Category: model refinement |
| CTF correction | Type: PHASE FLIPPING AND AMPLITUDE CORRECTION |
| 3D reconstruction | Resolution: 3.4 Å / Resolution method: FSC 0.143 CUT-OFF / Num. of particles: 126719 / Symmetry type: POINT |
 Movie
Movie Controller
Controller












 PDBj
PDBj





