[English] 日本語
 Yorodumi
Yorodumi- EMDB-7898: recombinant, shortened Plasmodium falciparum Circumsporozoite Pro... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-7898 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | recombinant, shortened Plasmodium falciparum Circumsporozoite Protein (CSP) in complex with immunoglobulin g (IgG) 311 . | |||||||||
 Map data Map data | Recombinant, shortened circumsporozoite protein in complex with immunoglobulin G 311 (IgG311) | |||||||||
 Sample Sample |
| |||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / negative staining / Resolution: 27.0 Å | |||||||||
 Authors Authors | Torres JL / Ward AB | |||||||||
 Citation Citation |  Journal: Sci Adv / Year: 2018 Journal: Sci Adv / Year: 2018Title: Cryo-EM structure of circumsporozoite protein with a vaccine-elicited antibody is stabilized by somatically mutated inter-Fab contacts. Authors: David Oyen / Jonathan L Torres / Christopher A Cottrell / C Richter King / Ian A Wilson / Andrew B Ward /  Abstract: The circumsporozoite protein (CSP) on the surface of sporozoites is important for parasite development, motility, and host hepatocyte invasion. However, intrinsic disorder of the NANP repeat ...The circumsporozoite protein (CSP) on the surface of sporozoites is important for parasite development, motility, and host hepatocyte invasion. However, intrinsic disorder of the NANP repeat sequence in the central region of CSP has hindered its structural and functional characterization. Here, the cryo-electron microscopy structure at ~3.4-Å resolution of a recombinant shortened CSP construct with the variable domains (Fabs) of a highly protective monoclonal antibody reveals an extended spiral conformation of the central NANP repeat region surrounded by antibodies. This unusual structure appears to be stabilized and/or induced by interaction with an antibody where contacts between adjacent Fabs are somatically mutated and enhance the interaction. This maturation in non-antigen contact residues may be an effective mechanism for antibodies to target tandem repeat sequences and provide novel insights into malaria vaccine design. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_7898.map.gz emd_7898.map.gz | 33.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-7898-v30.xml emd-7898-v30.xml emd-7898.xml emd-7898.xml | 10.8 KB 10.8 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_7898.png emd_7898.png | 45.5 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-7898 http://ftp.pdbj.org/pub/emdb/structures/EMD-7898 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7898 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7898 | HTTPS FTP |
-Validation report
| Summary document |  emd_7898_validation.pdf.gz emd_7898_validation.pdf.gz | 78.1 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_7898_full_validation.pdf.gz emd_7898_full_validation.pdf.gz | 77.2 KB | Display | |
| Data in XML |  emd_7898_validation.xml.gz emd_7898_validation.xml.gz | 494 B | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-7898 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-7898 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-7898 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-7898 | HTTPS FTP |
-Related structure data
| Related structure data |  7897C  7899C  9065C  9114C  6mb3C  6mhgC C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_7898.map.gz / Format: CCP4 / Size: 42.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_7898.map.gz / Format: CCP4 / Size: 42.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Recombinant, shortened circumsporozoite protein in complex with immunoglobulin G 311 (IgG311) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.05 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
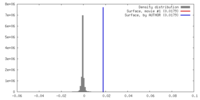
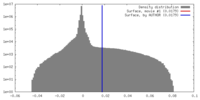
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Recombinant, shortened Plasmodium Falciparum Circumsprozoite Prot...
| Entire | Name: Recombinant, shortened Plasmodium Falciparum Circumsprozoite Protein (CSP) in complex with Immunoglobulin G (IgG) 311 |
|---|---|
| Components |
|
-Supramolecule #1: Recombinant, shortened Plasmodium Falciparum Circumsprozoite Prot...
| Supramolecule | Name: Recombinant, shortened Plasmodium Falciparum Circumsprozoite Protein (CSP) in complex with Immunoglobulin G (IgG) 311 type: complex / ID: 1 / Parent: 0 |
|---|
-Supramolecule #2: Recombinant, shortened Plasmodium Falciparum Circumsprozoite Prot...
| Supramolecule | Name: Recombinant, shortened Plasmodium Falciparum Circumsprozoite Protein (CSP) type: organelle_or_cellular_component / ID: 2 / Parent: 1 |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
-Supramolecule #3: Immunoglobulin G (IgG) 311
| Supramolecule | Name: Immunoglobulin G (IgG) 311 / type: organelle_or_cellular_component / ID: 3 / Parent: 1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  |
-Experimental details
-Structure determination
| Method | negative staining |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.01 mg/mL |
|---|---|
| Buffer | pH: 7.4 / Details: 1X TBS pH 7.4 |
| Staining | Type: NEGATIVE / Material: 2% Uranyl Formate Details: add 3 uL sample, blot, add 3 uL 2% uranyl formate, blot immediately, add 3 uL 2% uranyl formate, blot after 30 seconds |
| Grid | Model: Homemade / Material: COPPER / Mesh: 400 / Support film - Material: CARBON / Support film - topology: CONTINUOUS / Pretreatment - Type: PLASMA CLEANING |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI SPIRIT |
|---|---|
| Details | Gris screening performed on FEI Mogagni |
| Image recording | Film or detector model: TVIPS TEMCAM-F416 (4k x 4k) / Number grids imaged: 1 / Number real images: 199 / Average exposure time: 0.5 sec. / Average electron dose: 25.0 e/Å2 |
| Electron beam | Acceleration voltage: 120 kV / Electron source: LAB6 |
| Electron optics | Calibrated magnification: 52000 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm |
| Sample stage | Specimen holder model: SIDE ENTRY, EUCENTRIC / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Tecnai Spirit / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller


 UCSF Chimera
UCSF Chimera





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)