+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-7127 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of the cold- and menthol-sensing ion channel TRPM8 | |||||||||
 Map data Map data | Single-particle cryo-EM reconstruction of Transient Receptor Potential Melastatin channel TRPM8. | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | cold sensor / menthol sensor / calcium-permeable ion channel / ion channel / TRANSPORT PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationligand-gated calcium channel activity / identical protein binding / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Ficedula albicollis (Collared flycatcher) Ficedula albicollis (Collared flycatcher) | |||||||||
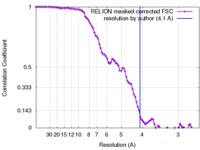
| Method | single particle reconstruction / cryo EM / Resolution: 4.1 Å | |||||||||
 Authors Authors | Yin Y / Wu M | |||||||||
 Citation Citation |  Journal: Science / Year: 2018 Journal: Science / Year: 2018Title: Structure of the cold- and menthol-sensing ion channel TRPM8. Authors: Ying Yin / Mengyu Wu / Lejla Zubcevic / William F Borschel / Gabriel C Lander / Seok-Yong Lee /  Abstract: Transient receptor potential melastatin (TRPM) cation channels are polymodal sensors that are involved in a variety of physiological processes. Within the TRPM family, member 8 (TRPM8) is the primary ...Transient receptor potential melastatin (TRPM) cation channels are polymodal sensors that are involved in a variety of physiological processes. Within the TRPM family, member 8 (TRPM8) is the primary cold and menthol sensor in humans. We determined the cryo-electron microscopy structure of the full-length TRPM8 from the collared flycatcher at an overall resolution of ~4.1 ångstroms. Our TRPM8 structure reveals a three-layered architecture. The amino-terminal domain with a fold distinct among known TRP structures, together with the carboxyl-terminal region, forms a large two-layered cytosolic ring that extensively interacts with the transmembrane channel layer. The structure suggests that the menthol-binding site is located within the voltage-sensor-like domain and thus provides a structural glimpse of the design principle of the molecular transducer for cold and menthol sensation. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_7127.map.gz emd_7127.map.gz | 59.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-7127-v30.xml emd-7127-v30.xml emd-7127.xml emd-7127.xml | 17.8 KB 17.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_7127_fsc.xml emd_7127_fsc.xml | 9.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_7127.png emd_7127.png | 196.9 KB | ||
| Filedesc metadata |  emd-7127.cif.gz emd-7127.cif.gz | 6.3 KB | ||
| Others |  emd_7127_half_map_1.map.gz emd_7127_half_map_1.map.gz emd_7127_half_map_2.map.gz emd_7127_half_map_2.map.gz | 45.8 MB 45.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-7127 http://ftp.pdbj.org/pub/emdb/structures/EMD-7127 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7127 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7127 | HTTPS FTP |
-Validation report
| Summary document |  emd_7127_validation.pdf.gz emd_7127_validation.pdf.gz | 1019.1 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_7127_full_validation.pdf.gz emd_7127_full_validation.pdf.gz | 1018.7 KB | Display | |
| Data in XML |  emd_7127_validation.xml.gz emd_7127_validation.xml.gz | 15.5 KB | Display | |
| Data in CIF |  emd_7127_validation.cif.gz emd_7127_validation.cif.gz | 20.6 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-7127 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-7127 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-7127 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-7127 | HTTPS FTP |
-Related structure data
| Related structure data |  6bpqMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_7127.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_7127.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Single-particle cryo-EM reconstruction of Transient Receptor Potential Melastatin channel TRPM8. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.31 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Half map: Odd half map
| File | emd_7127_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Odd half map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Even half map
| File | emd_7127_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Even half map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Transient receptor potential cation channel subfamily M member 8 ...
| Entire | Name: Transient receptor potential cation channel subfamily M member 8 (TRPM8) |
|---|---|
| Components |
|
-Supramolecule #1: Transient receptor potential cation channel subfamily M member 8 ...
| Supramolecule | Name: Transient receptor potential cation channel subfamily M member 8 (TRPM8) type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Ficedula albicollis (Collared flycatcher) Ficedula albicollis (Collared flycatcher) |
-Macromolecule #1: Transient receptor potential cation channel subfamily M member 8
| Macromolecule | Name: Transient receptor potential cation channel subfamily M member 8 type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Ficedula albicollis (Collared flycatcher) Ficedula albicollis (Collared flycatcher) |
| Molecular weight | Theoretical: 107.845281 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)CDTDSETLY DLMTQHWHLK TPNLVISVTG GAKNFALKPR MRKIFSRLIY IAQSKGAWIF T GGTHYGLM KYIGEVVRDN ...String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)CDTDSETLY DLMTQHWHLK TPNLVISVTG GAKNFALKPR MRKIFSRLIY IAQSKGAWIF T GGTHYGLM KYIGEVVRDN TISRSSEENV VAIGIA(UNK)(UNK)(UNK)(UNK) (UNK)LKRDPLYCL DNNHTHLLLV DN GTHGHPT IEAKVRTQLE KYISERVIPE SNYGGKIPIV CFAQGGGKET LKSINVAIKS KIPCVVVEGS GRIADVIASL VEA EGTLAS SCVKESLLRF LPRTISRLSE EETESWIKWI KEVLESPHLL TVIKIEEAGD EIVSNAISFA LYKAFSTNEH DRDN WNGQL KLLLEWNQLD LASDEIFTND RNWESADLQD VMFTALVKDR PKFVRLFLEN GLNLRKFLTT EVLRELYTNN FSSLV FKNL QIAKNSYNDA LLTFVWKMVE DFRRGAKRDD KNSKDEMEIE LSEECPITRH PLQALFIWSV LQNKKELSKV IWEQTR GCT LAALGASKLL KSMAKVKNDI NAAGESEELA NEYETRAVEL FTECYSNDED LAEQLLTYSC EAWGGSNCLE LAVEARD QQ FIAQPGVQNF LSKQWYGEIS RDTKNWKIIL CLFFFPLIGC GFISFRKKPV EKTKKLFLYY VSFFTSPFVV FSWNVIFY I AFLLLFAYVL LMDFQKEPTA LEIILYVLVF ILLCDEVRQW YMNGSKYFSD LWNVMDTLAI FYFIAGIVFR LHSDESSWY SGRVIFCLDY IVFTLRLIHI FTVSRNLGPK IIMLQRMMID VFFFLFLFAV WMVAFGVARQ GILRKNEHRW EWIFRSVIYE PYLAMFGQY PDDIDGTTYN FDHCTFSGNE SKPLCVELDA NNQPRFPEWI TIPLVCIYML STNILLVNLL VAMFGYTVGS V QENNDQVW KFQRFFLVQE YCSRLT(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)RNEDN EILAWEAVMK ENYLVKINT(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.5 mg/mL |
|---|---|
| Buffer | pH: 8 |
| Grid | Model: Quantifoil, UltrAuFoil, R1.2/1.3 / Material: GOLD / Mesh: 300 / Support film - Material: CARBON / Support film - topology: CONTINUOUS / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. / Details: 15 mA |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277.15 K / Instrument: HOMEMADE PLUNGER |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Dimensions - Width: 3710 pixel / Digitization - Dimensions - Height: 3838 pixel / Digitization - Frames/image: 1-48 / Number real images: 8367 / Average exposure time: 12.0 sec. / Average electron dose: 56.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.2 µm / Nominal magnification: 22500 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: RIGID BODY FIT / Overall B value: 96 |
|---|---|
| Output model |  PDB-6bpq: |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)