+ データを開く
データを開く
- 基本情報
基本情報
| 登録情報 | データベース: PDB / ID: 6qlf | ||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
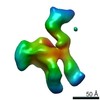
| タイトル | Structure of inner kinetochore CCAN complex with mask1 | ||||||||||||||||||||||||||||||||||||||||||
 要素 要素 | (Inner kinetochore subunit ...) x 8 | ||||||||||||||||||||||||||||||||||||||||||
 キーワード キーワード | DNA BINDING PROTEIN / inner kinetochore / CCAN | ||||||||||||||||||||||||||||||||||||||||||
| 機能・相同性 |  機能・相同性情報 機能・相同性情報negative regulation of meiotic DNA double-strand break formation involved in reciprocal meiotic recombination / COMA complex / maintenance of meiotic sister chromatid cohesion / Mis6-Sim4 complex / : / meiotic sister chromatid segregation / ascospore formation / establishment of meiotic sister chromatid cohesion / attachment of spindle microtubules to kinetochore / CENP-A containing chromatin assembly ...negative regulation of meiotic DNA double-strand break formation involved in reciprocal meiotic recombination / COMA complex / maintenance of meiotic sister chromatid cohesion / Mis6-Sim4 complex / : / meiotic sister chromatid segregation / ascospore formation / establishment of meiotic sister chromatid cohesion / attachment of spindle microtubules to kinetochore / CENP-A containing chromatin assembly / outer kinetochore / kinetochore assembly / protein localization to chromosome, centromeric region / establishment of mitotic sister chromatid cohesion / protein localization to kinetochore / spindle pole body / mitotic spindle assembly checkpoint signaling / meiotic cell cycle / chromosome segregation / kinetochore / cell division / structural molecule activity / nucleus / cytoplasm 類似検索 - 分子機能 | ||||||||||||||||||||||||||||||||||||||||||
| 生物種 |  | ||||||||||||||||||||||||||||||||||||||||||
| 手法 | 電子顕微鏡法 / 単粒子再構成法 / クライオ電子顕微鏡法 / 解像度: 3.45 Å | ||||||||||||||||||||||||||||||||||||||||||
 データ登録者 データ登録者 | Yan, K. / Yang, J. / Zhang, Z. / McLaughlin, S.H. / Chang, L. / Fasci, D. / Heck, A.J.R. / Barford, D. | ||||||||||||||||||||||||||||||||||||||||||
| 資金援助 |  英国, 3件 英国, 3件
| ||||||||||||||||||||||||||||||||||||||||||
 引用 引用 |  ジャーナル: Nature / 年: 2019 ジャーナル: Nature / 年: 2019タイトル: Structure of the inner kinetochore CCAN complex assembled onto a centromeric nucleosome. 著者: Kaige Yan / Jing Yang / Ziguo Zhang / Stephen H McLaughlin / Leifu Chang / Domenico Fasci / Ann E Ehrenhofer-Murray / Albert J R Heck / David Barford /     要旨: In eukaryotes, accurate chromosome segregation in mitosis and meiosis maintains genome stability and prevents aneuploidy. Kinetochores are large protein complexes that, by assembling onto specialized ...In eukaryotes, accurate chromosome segregation in mitosis and meiosis maintains genome stability and prevents aneuploidy. Kinetochores are large protein complexes that, by assembling onto specialized Cenp-A nucleosomes, function to connect centromeric chromatin to microtubules of the mitotic spindle. Whereas the centromeres of vertebrate chromosomes comprise millions of DNA base pairs and attach to multiple microtubules, the simple point centromeres of budding yeast are connected to individual microtubules. All 16 budding yeast chromosomes assemble complete kinetochores using a single Cenp-A nucleosome (Cenp-A), each of which is perfectly centred on its cognate centromere. The inner and outer kinetochore modules are responsible for interacting with centromeric chromatin and microtubules, respectively. Here we describe the cryo-electron microscopy structure of the Saccharomyces cerevisiae inner kinetochore module, the constitutive centromere associated network (CCAN) complex, assembled onto a Cenp-A nucleosome (CCAN-Cenp-A). The structure explains the interdependency of the constituent subcomplexes of CCAN and shows how the Y-shaped opening of CCAN accommodates Cenp-A to enable specific CCAN subunits to contact the nucleosomal DNA and histone subunits. Interactions with the unwrapped DNA duplex at the two termini of Cenp-A are mediated predominantly by a DNA-binding groove in the Cenp-L-Cenp-N subcomplex. Disruption of these interactions impairs assembly of CCAN onto Cenp-A. Our data indicate a mechanism of Cenp-A nucleosome recognition by CCAN and how CCAN acts as a platform for assembly of the outer kinetochore to link centromeres to the mitotic spindle for chromosome segregation. | ||||||||||||||||||||||||||||||||||||||||||
| 履歴 |
|
- 構造の表示
構造の表示
| ムービー |
 ムービービューア ムービービューア |
|---|---|
| 構造ビューア | 分子:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
- ダウンロードとリンク
ダウンロードとリンク
- ダウンロード
ダウンロード
| PDBx/mmCIF形式 |  6qlf.cif.gz 6qlf.cif.gz | 334 KB | 表示 |  PDBx/mmCIF形式 PDBx/mmCIF形式 |
|---|---|---|---|---|
| PDB形式 |  pdb6qlf.ent.gz pdb6qlf.ent.gz | 254.1 KB | 表示 |  PDB形式 PDB形式 |
| PDBx/mmJSON形式 |  6qlf.json.gz 6qlf.json.gz | ツリー表示 |  PDBx/mmJSON形式 PDBx/mmJSON形式 | |
| その他 |  その他のダウンロード その他のダウンロード |
-検証レポート
| 文書・要旨 |  6qlf_validation.pdf.gz 6qlf_validation.pdf.gz | 727.3 KB | 表示 |  wwPDB検証レポート wwPDB検証レポート |
|---|---|---|---|---|
| 文書・詳細版 |  6qlf_full_validation.pdf.gz 6qlf_full_validation.pdf.gz | 735.7 KB | 表示 | |
| XML形式データ |  6qlf_validation.xml.gz 6qlf_validation.xml.gz | 46.8 KB | 表示 | |
| CIF形式データ |  6qlf_validation.cif.gz 6qlf_validation.cif.gz | 74.3 KB | 表示 | |
| アーカイブディレクトリ |  https://data.pdbj.org/pub/pdb/validation_reports/ql/6qlf https://data.pdbj.org/pub/pdb/validation_reports/ql/6qlf ftp://data.pdbj.org/pub/pdb/validation_reports/ql/6qlf ftp://data.pdbj.org/pub/pdb/validation_reports/ql/6qlf | HTTPS FTP |
-関連構造データ
- リンク
リンク
- 集合体
集合体
| 登録構造単位 | 
|
|---|---|
| 1 |
|
- 要素
要素
-Inner kinetochore subunit ... , 8種, 8分子 LNOPQUYZ
| #1: タンパク質 | 分子量: 28093.223 Da / 分子数: 1 / 由来タイプ: 組換発現 由来: (組換発現)  遺伝子: IML3, MCM19, YBR107C, YBR0836 / 細胞株 (発現宿主): High Five / 発現宿主:  Trichoplusia ni (イラクサキンウワバ) / 参照: UniProt: P38265 Trichoplusia ni (イラクサキンウワバ) / 参照: UniProt: P38265 |
|---|---|
| #2: タンパク質 | 分子量: 53538.566 Da / 分子数: 1 / 由来タイプ: 組換発現 由来: (組換発現)  遺伝子: CHL4, CTF17, MCM17, YDR254W, YD9320A.04 / 細胞株 (発現宿主): High Five / 発現宿主:  Trichoplusia ni (イラクサキンウワバ) / 参照: UniProt: P38907 Trichoplusia ni (イラクサキンウワバ) / 参照: UniProt: P38907 |
| #3: タンパク質 | 分子量: 43028.879 Da / 分子数: 1 / 由来タイプ: 組換発現 由来: (組換発現)  遺伝子: MCM21, CTF5, YDR318W / 細胞株 (発現宿主): High Five / 発現宿主:  Trichoplusia ni (イラクサキンウワバ) / 参照: UniProt: Q06675 Trichoplusia ni (イラクサキンウワバ) / 参照: UniProt: Q06675 |
| #4: タンパク質 | 分子量: 42841.113 Da / 分子数: 1 / 由来タイプ: 組換発現 由来: (組換発現)  遺伝子: CTF19, MCM18, YPL018W, LPB13W / 細胞株 (発現宿主): High Five / 発現宿主:  Trichoplusia ni (イラクサキンウワバ) / 参照: UniProt: Q02732 Trichoplusia ni (イラクサキンウワバ) / 参照: UniProt: Q02732 |
| #5: タンパク質 | 分子量: 47427.246 Da / 分子数: 1 / 由来タイプ: 組換発現 由来: (組換発現)  遺伝子: OKP1, YGR179C / 細胞株 (発現宿主): High Five / 発現宿主:  Trichoplusia ni (イラクサキンウワバ) / 参照: UniProt: P53298 Trichoplusia ni (イラクサキンウワバ) / 参照: UniProt: P53298 |
| #6: タンパク質 | 分子量: 37081.246 Da / 分子数: 1 / 由来タイプ: 組換発現 由来: (組換発現)  遺伝子: AME1, ARP100, YBR211C, YBR1458 / 細胞株 (発現宿主): High Five / 発現宿主:  Trichoplusia ni (イラクサキンウワバ) / 参照: UniProt: P38313 Trichoplusia ni (イラクサキンウワバ) / 参照: UniProt: P38313 |
| #7: タンパク質 | 分子量: 27006.451 Da / 分子数: 1 / 由来タイプ: 組換発現 由来: (組換発現)  遺伝子: NKP1, YDR383C / 細胞株 (発現宿主): High Five / 発現宿主:  Trichoplusia ni (イラクサキンウワバ) / 参照: UniProt: Q12493 Trichoplusia ni (イラクサキンウワバ) / 参照: UniProt: Q12493 |
| #8: タンパク質 | 分子量: 17877.033 Da / 分子数: 1 / 由来タイプ: 組換発現 由来: (組換発現)  遺伝子: NKP2, YLR315W / 細胞株 (発現宿主): High Five / 発現宿主:  Trichoplusia ni (イラクサキンウワバ) / 参照: UniProt: Q06162 Trichoplusia ni (イラクサキンウワバ) / 参照: UniProt: Q06162 |
-詳細
| Has protein modification | N |
|---|
-実験情報
-実験
| 実験 | 手法: 電子顕微鏡法 |
|---|---|
| EM実験 | 試料の集合状態: PARTICLE / 3次元再構成法: 単粒子再構成法 |
- 試料調製
試料調製
| 構成要素 | 名称: Inner kinetochore CCAN complex / タイプ: COMPLEX / Entity ID: all / 由来: RECOMBINANT |
|---|---|
| 由来(天然) | 生物種:  |
| 由来(組換発現) | 生物種:  Trichoplusia ni (イラクサキンウワバ) Trichoplusia ni (イラクサキンウワバ) |
| 緩衝液 | pH: 8 |
| 試料 | 濃度: 1 mg/ml / 包埋: NO / シャドウイング: NO / 染色: NO / 凍結: YES |
| 急速凍結 | 凍結剤: ETHANE |
- 電子顕微鏡撮影
電子顕微鏡撮影
| 実験機器 |  モデル: Titan Krios / 画像提供: FEI Company |
|---|---|
| 顕微鏡 | モデル: FEI TITAN KRIOS |
| 電子銃 | 電子線源:  FIELD EMISSION GUN / 加速電圧: 300 kV / 照射モード: FLOOD BEAM FIELD EMISSION GUN / 加速電圧: 300 kV / 照射モード: FLOOD BEAM |
| 電子レンズ | モード: BRIGHT FIELD |
| 撮影 | 電子線照射量: 32 e/Å2 / 検出モード: COUNTING フィルム・検出器のモデル: FEI FALCON III (4k x 4k) |
- 解析
解析
| ソフトウェア | 名称: PHENIX / バージョン: 1.14_3260: / 分類: 精密化 |
|---|---|
| EMソフトウェア | 名称: PHENIX / カテゴリ: モデル精密化 |
| CTF補正 | タイプ: PHASE FLIPPING AND AMPLITUDE CORRECTION |
| 3次元再構成 | 解像度: 3.45 Å / 解像度の算出法: FSC 0.143 CUT-OFF / 粒子像の数: 465029 / 対称性のタイプ: POINT |
 ムービー
ムービー コントローラー
コントローラー
















 PDBj
PDBj