登録情報 データベース : PDB / ID : 6v79タイトル Crystal structure of Danio rerio histone deacetylase 6 catalytic domain 2 (CD2) complexed with NF2376 Hdac6 protein キーワード / 機能・相同性 分子機能 ドメイン・相同性 構成要素
/ / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / 生物種 Danio rerio (ゼブラフィッシュ)手法 / / / 解像度 : 2.03951514036 Å データ登録者 Osko, J.D. / Christianson, D.W. 資金援助 組織 認可番号 国 National Institutes of Health/National Institute of General Medical Sciences (NIH/NIGMS) GM49758
ジャーナル : J.Med.Chem. / 年 : 2021タイトル : Harnessing the Role of HDAC6 in Idiopathic Pulmonary Fibrosis: Design, Synthesis, Structural Analysis, and Biological Evaluation of Potent Inhibitors.著者: Campiani, G. / Cavella, C. / Osko, J.D. / Brindisi, M. / Relitti, N. / Brogi, S. / Saraswati, A.P. / Federico, S. / Chemi, G. / Maramai, S. / Carullo, G. / Jaeger, B. / Carleo, A. / ... 著者 : Campiani, G. / Cavella, C. / Osko, J.D. / Brindisi, M. / Relitti, N. / Brogi, S. / Saraswati, A.P. / Federico, S. / Chemi, G. / Maramai, S. / Carullo, G. / Jaeger, B. / Carleo, A. / Benedetti, R. / Sarno, F. / Lamponi, S. / Rottoli, P. / Bargagli, E. / Bertucci, C. / Tedesco, D. / Herp, D. / Senger, J. / Ruberti, G. / Saccoccia, F. / Saponara, S. / Gorelli, B. / Valoti, M. / Kennedy, B. / Sundaramurthi, H. / Butini, S. / Jung, M. / Roach, K.M. / Altucci, L. / Bradding, P. / Christianson, D.W. / Gemma, S. / Prasse, A. 履歴 登録 2019年12月8日 登録サイト / 処理サイト 改定 1.0 2020年12月2日 Provider / タイプ 改定 1.1 2021年7月28日 Group / カテゴリ / citation_authorItem _citation.country / _citation.journal_abbrev ... _citation.country / _citation.journal_abbrev / _citation.journal_id_ASTM / _citation.journal_id_CSD / _citation.journal_id_ISSN / _citation.pdbx_database_id_DOI / _citation.pdbx_database_id_PubMed / _citation.title / _citation.year 改定 1.2 2021年8月4日 Group / カテゴリ Item / _citation.page_first / _citation.page_last改定 1.3 2023年10月11日 Group Advisory / Data collection ... Advisory / Data collection / Database references / Refinement description カテゴリ chem_comp_atom / chem_comp_bond ... chem_comp_atom / chem_comp_bond / database_2 / pdbx_initial_refinement_model / pdbx_unobs_or_zero_occ_atoms Item / _database_2.pdbx_database_accession
すべて表示 表示を減らす
 データを開く
データを開く 基本情報
基本情報 要素
要素 キーワード
キーワード 機能・相同性情報
機能・相同性情報
 X線回折 /
X線回折 /  シンクロトロン /
シンクロトロン /  分子置換 / 解像度: 2.03951514036 Å
分子置換 / 解像度: 2.03951514036 Å  データ登録者
データ登録者 米国, 1件
米国, 1件  引用
引用 ジャーナル: J.Med.Chem. / 年: 2021
ジャーナル: J.Med.Chem. / 年: 2021 構造の表示
構造の表示 Molmil
Molmil Jmol/JSmol
Jmol/JSmol ダウンロードとリンク
ダウンロードとリンク ダウンロード
ダウンロード 6v79.cif.gz
6v79.cif.gz PDBx/mmCIF形式
PDBx/mmCIF形式 pdb6v79.ent.gz
pdb6v79.ent.gz PDB形式
PDB形式 6v79.json.gz
6v79.json.gz PDBx/mmJSON形式
PDBx/mmJSON形式 その他のダウンロード
その他のダウンロード 6v79_validation.pdf.gz
6v79_validation.pdf.gz wwPDB検証レポート
wwPDB検証レポート 6v79_full_validation.pdf.gz
6v79_full_validation.pdf.gz 6v79_validation.xml.gz
6v79_validation.xml.gz 6v79_validation.cif.gz
6v79_validation.cif.gz https://data.pdbj.org/pub/pdb/validation_reports/v7/6v79
https://data.pdbj.org/pub/pdb/validation_reports/v7/6v79 ftp://data.pdbj.org/pub/pdb/validation_reports/v7/6v79
ftp://data.pdbj.org/pub/pdb/validation_reports/v7/6v79
 リンク
リンク 集合体
集合体


 要素
要素



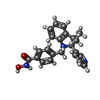






 X線回折 / 使用した結晶の数: 1
X線回折 / 使用した結晶の数: 1  試料調製
試料調製 シンクロトロン / サイト:
シンクロトロン / サイト:  APS
APS  / ビームライン: 24-ID-C / 波長: 0.98 Å
/ ビームライン: 24-ID-C / 波長: 0.98 Å 解析
解析 分子置換
分子置換 ムービー
ムービー コントローラー
コントローラー
















 PDBj
PDBj



