+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 6v0p | ||||||
|---|---|---|---|---|---|---|---|
| Title | PRMT5 complex bound to covalent PBM inhibitor BRD6711 | ||||||
 Components Components |
| ||||||
 Keywords Keywords | SPLICING/TRANSFERASE / methyltransferase / splicing / SDMA / epigenetic / SPLICING-TRANSFERASE complex | ||||||
| Function / homology |  Function and homology information Function and homology informationpositive regulation of adenylate cyclase-inhibiting dopamine receptor signaling pathway / peptidyl-arginine N-methylation / oocyte axis specification / type II protein arginine methyltransferase / protein-arginine omega-N symmetric methyltransferase activity / peptidyl-arginine methylation / Golgi ribbon formation / negative regulation of epithelial cell proliferation involved in prostate gland development / secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development / histone H4R3 methyltransferase activity ...positive regulation of adenylate cyclase-inhibiting dopamine receptor signaling pathway / peptidyl-arginine N-methylation / oocyte axis specification / type II protein arginine methyltransferase / protein-arginine omega-N symmetric methyltransferase activity / peptidyl-arginine methylation / Golgi ribbon formation / negative regulation of epithelial cell proliferation involved in prostate gland development / secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development / histone H4R3 methyltransferase activity / : / epithelial cell proliferation involved in prostate gland development / protein-arginine N-methyltransferase activity / methylosome / positive regulation of mRNA splicing, via spliceosome / methyl-CpG binding / endothelial cell activation / histone H3 methyltransferase activity / histone methyltransferase activity / regulation of mitotic nuclear division / negative regulation of gene expression via chromosomal CpG island methylation / Cul4B-RING E3 ubiquitin ligase complex / E-box binding / histone methyltransferase complex / positive regulation of oligodendrocyte differentiation / negative regulation of cell differentiation / spliceosomal snRNP assembly / ribonucleoprotein complex binding / regulation of ERK1 and ERK2 cascade / ubiquitin-like ligase-substrate adaptor activity / liver regeneration / regulation of signal transduction by p53 class mediator / methyltransferase activity / circadian regulation of gene expression / DNA-templated transcription termination / Regulation of TP53 Activity through Methylation / RMTs methylate histone arginines / protein polyubiquitination / p53 binding / transcription corepressor activity / snRNP Assembly / ubiquitin-dependent protein catabolic process / transcription coactivator activity / chromatin remodeling / protein heterodimerization activity / positive regulation of cell population proliferation / regulation of DNA-templated transcription / regulation of transcription by RNA polymerase II / chromatin / Golgi apparatus / nucleoplasm / identical protein binding / nucleus / cytosol / cytoplasm Similarity search - Function | ||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 1.88 Å MOLECULAR REPLACEMENT / Resolution: 1.88 Å | ||||||
 Authors Authors | McMillan, B.J. / McKinney, D.C. | ||||||
 Citation Citation |  Journal: J Med Chem / Year: 2021 Journal: J Med Chem / Year: 2021Title: Discovery of a First-in-Class Inhibitor of the PRMT5-Substrate Adaptor Interaction. Authors: David C McKinney / Brian J McMillan / Matthew J Ranaghan / Jamie A Moroco / Merissa Brousseau / Zachary Mullin-Bernstein / Meghan O'Keefe / Patrick McCarren / Michael F Mesleh / Kathleen M ...Authors: David C McKinney / Brian J McMillan / Matthew J Ranaghan / Jamie A Moroco / Merissa Brousseau / Zachary Mullin-Bernstein / Meghan O'Keefe / Patrick McCarren / Michael F Mesleh / Kathleen M Mulvaney / Foxy Robinson / Ritu Singh / Besnik Bajrami / Florence F Wagner / Robert Hilgraf / Martin J Drysdale / Arthur J Campbell / Adam Skepner / David E Timm / Dale Porter / Virendar K Kaushik / William R Sellers / Alessandra Ianari /  Abstract: PRMT5 and its substrate adaptor proteins (SAPs), pICln and Riok1, are synthetic lethal dependencies in MTAP-deleted cancer cells. SAPs share a conserved PRMT5 binding motif (PBM) which mediates ...PRMT5 and its substrate adaptor proteins (SAPs), pICln and Riok1, are synthetic lethal dependencies in MTAP-deleted cancer cells. SAPs share a conserved PRMT5 binding motif (PBM) which mediates binding to a surface of PRMT5 distal to the catalytic site. This interaction is required for methylation of several PRMT5 substrates, including histone and spliceosome complexes. We screened for small molecule inhibitors of the PRMT5-PBM interaction and validated a compound series which binds to the PRMT5-PBM interface and directly inhibits binding of SAPs. Mode of action studies revealed the formation of a covalent bond between a halogenated pyridazinone group and cysteine 278 of PRMT5. Optimization of the starting hit produced a lead compound, BRD0639, which engages the target in cells, disrupts PRMT5-RIOK1 complexes, and reduces substrate methylation. BRD0639 is a first-in-class PBM-competitive inhibitor that can support studies of PBM-dependent PRMT5 activities and the development of novel PRMT5 inhibitors that selectively target these functions. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  6v0p.cif.gz 6v0p.cif.gz | 753.4 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb6v0p.ent.gz pdb6v0p.ent.gz | 601 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  6v0p.json.gz 6v0p.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/v0/6v0p https://data.pdbj.org/pub/pdb/validation_reports/v0/6v0p ftp://data.pdbj.org/pub/pdb/validation_reports/v0/6v0p ftp://data.pdbj.org/pub/pdb/validation_reports/v0/6v0p | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  7m05C  4gqbS S: Starting model for refinement C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||||
| Unit cell |
|
- Components
Components
-Protein , 2 types, 2 molecules AB
| #1: Protein | Mass: 72766.664 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: PRMT5, HRMT1L5, IBP72, JBP1, SKB1 / Production host: Homo sapiens (human) / Gene: PRMT5, HRMT1L5, IBP72, JBP1, SKB1 / Production host:  References: UniProt: O14744, type II protein arginine methyltransferase |
|---|---|
| #2: Protein | Mass: 36723.164 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: WDR77, MEP50, WD45, HKMT1069, Nbla10071 / Production host: Homo sapiens (human) / Gene: WDR77, MEP50, WD45, HKMT1069, Nbla10071 / Production host:  |
-Non-polymers , 5 types, 643 molecules 

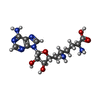






| #3: Chemical | ChemComp-QN4 / | ||||||
|---|---|---|---|---|---|---|---|
| #4: Chemical | | #5: Chemical | ChemComp-SFG / | #6: Chemical | #7: Water | ChemComp-HOH / | |
-Details
| Has ligand of interest | Y |
|---|---|
| Has protein modification | Y |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.8 Å3/Da / Density % sol: 56.03 % |
|---|---|
| Crystal grow | Temperature: 281 K / Method: vapor diffusion, hanging drop / pH: 7.4 / Details: 26% PEG3350 200 mM ammonium sulfate 0.05% w/v DDM |
-Data collection
| Diffraction | Mean temperature: 100 K / Serial crystal experiment: N |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  ESRF ESRF  / Beamline: ID30B / Wavelength: 0.979 Å / Beamline: ID30B / Wavelength: 0.979 Å |
| Detector | Type: DECTRIS PILATUS3 6M / Detector: PIXEL / Date: Jul 26, 2018 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 0.979 Å / Relative weight: 1 |
| Reflection | Resolution: 1.88→45 Å / Num. obs: 50976 / % possible obs: 93.9 % / Redundancy: 6.5 % / Biso Wilson estimate: 39.59 Å2 / CC1/2: 0.998 / Rrim(I) all: 0.115 / Net I/σ(I): 32.7 |
| Reflection shell | Resolution: 1.88→2.1 Å / Redundancy: 5.5 % / Mean I/σ(I) obs: 1.1 / Num. unique obs: 1020 / CC1/2: 0.48 / Rrim(I) all: 1.6 / % possible all: 86 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: 4GQB Resolution: 1.88→36.73 Å / Cross valid method: THROUGHOUT
| ||||||||||||||||||||||||
| Displacement parameters | Biso mean: 45.03 Å2 | ||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.88→36.73 Å
| ||||||||||||||||||||||||
| Refine LS restraints |
|
 Movie
Movie Controller
Controller













 PDBj
PDBj





