[English] 日本語
 Yorodumi
Yorodumi- PDB-6qhb: Crystal Structure of Human Kallikrein 6 in complex with GSK578724A -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 6qhb | ||||||
|---|---|---|---|---|---|---|---|
| Title | Crystal Structure of Human Kallikrein 6 in complex with GSK578724A | ||||||
 Components Components | Kallikrein-6 | ||||||
 Keywords Keywords | HYDROLASE / Protease / Inhibitor / Complex | ||||||
| Function / homology |  Function and homology information Function and homology informationcornified envelope / tissue regeneration / positive regulation of G protein-coupled receptor signaling pathway / amyloid precursor protein metabolic process / hormone metabolic process / regulation of neuron projection development / protein autoprocessing / regulation of cell differentiation / collagen catabolic process / Hydrolases; Acting on peptide bonds (peptidases); Serine endopeptidases ...cornified envelope / tissue regeneration / positive regulation of G protein-coupled receptor signaling pathway / amyloid precursor protein metabolic process / hormone metabolic process / regulation of neuron projection development / protein autoprocessing / regulation of cell differentiation / collagen catabolic process / Hydrolases; Acting on peptide bonds (peptidases); Serine endopeptidases / intercellular bridge / myelination / secretory granule / protein maturation / central nervous system development / response to wounding / nuclear membrane / serine-type endopeptidase activity / nucleolus / endoplasmic reticulum / mitochondrion / extracellular space / extracellular region / nucleoplasm / cytoplasm Similarity search - Function | ||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 1.84 Å MOLECULAR REPLACEMENT / Resolution: 1.84 Å | ||||||
 Authors Authors | Thorpe, J.H. | ||||||
 Citation Citation |  Journal: Bioorg. Med. Chem. Lett. / Year: 2019 Journal: Bioorg. Med. Chem. Lett. / Year: 2019Title: Kallikrein 5 inhibitors identified through structure based drug design in search for a treatment for Netherton Syndrome. Authors: White, G.V. / Edgar, E.V. / Holmes, D.S. / Lewell, X.Q. / Liddle, J. / Polyakova, O. / Smith, K.J. / Thorpe, J.H. / Walker, A.L. / Wang, Y. / Young, R.J. / Hovnanian, A. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  6qhb.cif.gz 6qhb.cif.gz | 108.4 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb6qhb.ent.gz pdb6qhb.ent.gz | 81.4 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  6qhb.json.gz 6qhb.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/qh/6qhb https://data.pdbj.org/pub/pdb/validation_reports/qh/6qhb ftp://data.pdbj.org/pub/pdb/validation_reports/qh/6qhb ftp://data.pdbj.org/pub/pdb/validation_reports/qh/6qhb | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  6qh9C  6qhaC  6qhcC  1lo6S S: Starting model for refinement C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 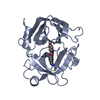
| ||||||||
| 2 | 
| ||||||||
| Unit cell |
|
- Components
Components
| #1: Protein | Mass: 24417.695 Da / Num. of mol.: 2 / Mutation: R74G/R76Q/N132Q Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: KLK6, PRSS18, PRSS9 / Production host: Homo sapiens (human) / Gene: KLK6, PRSS18, PRSS9 / Production host:  Baculovirus expression vector pFastBac1-HM Baculovirus expression vector pFastBac1-HMReferences: UniProt: Q92876, Hydrolases; Acting on peptide bonds (peptidases); Serine endopeptidases #2: Chemical | #3: Chemical | ChemComp-GOL / | #4: Water | ChemComp-HOH / | Has protein modification | Y | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.46 Å3/Da / Density % sol: 50.08 % |
|---|---|
| Crystal grow | Temperature: 293 K / Method: vapor diffusion, sitting drop / pH: 8.6 / Details: 100mM Tris-HCl pH 8.6, PEG 4000 18-28% |
-Data collection
| Diffraction | Mean temperature: 100 K / Serial crystal experiment: N |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  Diamond Diamond  / Beamline: I02 / Wavelength: 0.9795 Å / Beamline: I02 / Wavelength: 0.9795 Å |
| Detector | Type: DECTRIS PILATUS 6M / Detector: PIXEL / Date: Oct 25, 2013 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 0.9795 Å / Relative weight: 1 |
| Reflection | Resolution: 1.84→45.95 Å / Num. obs: 40448 / % possible obs: 97.2 % / Redundancy: 3.1 % / Biso Wilson estimate: 28.57 Å2 / CC1/2: 0.997 / Rmerge(I) obs: 0.063 / Rpim(I) all: 0.042 / Rrim(I) all: 0.076 / Net I/σ(I): 12.4 |
| Reflection shell | Resolution: 1.84→1.89 Å / Redundancy: 2.2 % / Rmerge(I) obs: 0.493 / Num. unique obs: 6129 / CC1/2: 0.599 / Rpim(I) all: 0.399 / Rrim(I) all: 0.638 / % possible all: 93.5 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: 1LO6 Resolution: 1.84→26.94 Å / Cor.coef. Fo:Fc: 0.956 / Cor.coef. Fo:Fc free: 0.949 / SU R Cruickshank DPI: 0.132 / Cross valid method: THROUGHOUT / σ(F): 0 / SU R Blow DPI: 0.139 / SU Rfree Blow DPI: 0.12 / SU Rfree Cruickshank DPI: 0.117
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 30.68 Å2
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine analyze | Luzzati coordinate error obs: 0.24 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: 1 / Resolution: 1.84→26.94 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 1.84→1.89 Å / Rfactor Rfree error: 0 / Total num. of bins used: 20
|
 Movie
Movie Controller
Controller











 PDBj
PDBj