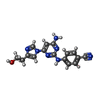
Entry Database : PDB / ID : 6m9hTitle JAK2 JH2 in complex with diaminopyrimidine JAK040 Tyrosine-protein kinase JAK2 Keywords / / Function / homology Function Domain/homology Component
/ / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / Biological species Homo sapiens (human)Method / / Resolution : 1.79 Å Authors Puleo, D.E. / Schlessinger, J. / Jorgensen, W.L. Funding support Organization Grant number Country National Institutes of Health/National Institute of General Medical Sciences (NIH/NIGMS) GM007324 Other private YG-003-13 National Institutes of Health/National Institute of General Medical Sciences (NIH/NIGMS) GM32136
Journal : To Be Published Title : JAK2 JH2 BindersAuthors : Puleo, D.E. / Schlessinger, J. History Deposition Aug 23, 2018 Deposition site / Processing site Revision 1.0 Sep 5, 2018 Provider / Type Revision 1.1 Nov 21, 2018 Group / Data collection / Structure summaryCategory / pdbx_audit_supportRevision 1.2 Jan 1, 2020 Group / Category / Item Revision 1.3 Mar 13, 2024 Group / Database references / Category / chem_comp_bond / database_2Item / _database_2.pdbx_database_accession
Show all Show less
 Open data
Open data Basic information
Basic information Components
Components Keywords
Keywords Function and homology information
Function and homology information Homo sapiens (human)
Homo sapiens (human) X-RAY DIFFRACTION /
X-RAY DIFFRACTION /  SYNCHROTRON / Resolution: 1.79 Å
SYNCHROTRON / Resolution: 1.79 Å  Authors
Authors United States, 3items
United States, 3items  Citation
Citation Journal: To Be Published
Journal: To Be Published Structure visualization
Structure visualization Molmil
Molmil Jmol/JSmol
Jmol/JSmol Downloads & links
Downloads & links Download
Download 6m9h.cif.gz
6m9h.cif.gz PDBx/mmCIF format
PDBx/mmCIF format pdb6m9h.ent.gz
pdb6m9h.ent.gz PDB format
PDB format 6m9h.json.gz
6m9h.json.gz PDBx/mmJSON format
PDBx/mmJSON format Other downloads
Other downloads https://data.pdbj.org/pub/pdb/validation_reports/m9/6m9h
https://data.pdbj.org/pub/pdb/validation_reports/m9/6m9h ftp://data.pdbj.org/pub/pdb/validation_reports/m9/6m9h
ftp://data.pdbj.org/pub/pdb/validation_reports/m9/6m9h Links
Links Assembly
Assembly
 Components
Components Homo sapiens (human) / Gene: JAK2 / Production host:
Homo sapiens (human) / Gene: JAK2 / Production host: 
 X-RAY DIFFRACTION / Number of used crystals: 1
X-RAY DIFFRACTION / Number of used crystals: 1  Sample preparation
Sample preparation SYNCHROTRON / Site:
SYNCHROTRON / Site:  APS
APS  / Beamline: 24-ID-E / Wavelength: 0.98 Å
/ Beamline: 24-ID-E / Wavelength: 0.98 Å Processing
Processing Movie
Movie Controller
Controller

















 PDBj
PDBj