[English] 日本語
 Yorodumi
Yorodumi- EMDB-6766: Local map for the Ha region of the phycobilisome from the red alg... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-6766 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Local map for the Ha region of the phycobilisome from the red alga Griffithsia pacifica | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Biological species |  Griffithsia pacifica (eukaryote) Griffithsia pacifica (eukaryote) | |||||||||
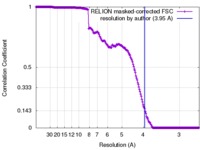
| Method | single particle reconstruction / cryo EM / Resolution: 3.95 Å | |||||||||
 Authors Authors | Zhang J / Ma JF / Sun S / Sui SF | |||||||||
 Citation Citation |  Journal: Nature / Year: 2017 Journal: Nature / Year: 2017Title: Structure of phycobilisome from the red alga Griffithsia pacifica. Authors: Jun Zhang / Jianfei Ma / Desheng Liu / Song Qin / Shan Sun / Jindong Zhao / Sen-Fang Sui /  Abstract: Life on Earth depends on photosynthesis for its conversion of solar energy to chemical energy. Photosynthetic organisms have developed a variety of light-harvesting systems to capture sunlight. The ...Life on Earth depends on photosynthesis for its conversion of solar energy to chemical energy. Photosynthetic organisms have developed a variety of light-harvesting systems to capture sunlight. The largest light-harvesting complex is the phycobilisome (PBS), the main light-harvesting antenna in cyanobacteria and red algae. It is composed of phycobiliproteins and linker proteins but the assembly mechanisms and energy transfer pathways of the PBS are not well understood. Here we report the structure of a 16.8-megadalton PBS from a red alga at 3.5 Å resolution obtained by single-particle cryo-electron microscopy. We modelled 862 protein subunits, including 4 linkers in the core, 16 rod-core linkers and 52 rod linkers, and located a total of 2,048 chromophores. This structure reveals the mechanisms underlying specific interactions between linkers and phycobiliproteins, and the formation of linker skeletons. These results provide a firm structural basis for our understanding of complex assembly and the mechanisms of energy transfer within the PBS. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_6766.map.gz emd_6766.map.gz | 625.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-6766-v30.xml emd-6766-v30.xml emd-6766.xml emd-6766.xml | 9.2 KB 9.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_6766_fsc.xml emd_6766_fsc.xml | 19.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_6766.png emd_6766.png | 313.2 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-6766 http://ftp.pdbj.org/pub/emdb/structures/EMD-6766 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6766 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6766 | HTTPS FTP |
-Related structure data
| Related structure data |  6758C  6759C  6760C  6761C  6762C  6763C  6764C  6765C  6767C  6768C  6769C  5y6pC C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_6766.map.gz / Format: CCP4 / Size: 669.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_6766.map.gz / Format: CCP4 / Size: 669.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.32 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : phycobilisome from the red alga Griffithsia pacifica
| Entire | Name: phycobilisome from the red alga Griffithsia pacifica |
|---|---|
| Components |
|
-Supramolecule #1: phycobilisome from the red alga Griffithsia pacifica
| Supramolecule | Name: phycobilisome from the red alga Griffithsia pacifica / type: complex / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  Griffithsia pacifica (eukaryote) Griffithsia pacifica (eukaryote) |
| Molecular weight | Experimental: 16.8 MDa |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1 mg/mL |
|---|---|
| Buffer | pH: 7 |
| Grid | Model: Qiagen / Material: COPPER / Mesh: 400 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Atmosphere: AIR / Details: The grid was coated with holy carbon |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 289.15 K |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Average electron dose: 8.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Calibrated defocus max: 2.3 µm / Calibrated defocus min: 1.4 µm / Calibrated magnification: 22500 / Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.3 µm / Nominal defocus min: 1.4 µm / Nominal magnification: 22500 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Protocol: AB INITIO MODEL |
|---|
 Movie
Movie Controller
Controller




 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)