[English] 日本語
 Yorodumi
Yorodumi- EMDB-6555: 2.9 Angstrom Resolution Cryo-EM 3-D Reconstruction of Close-packe... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-6555 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | 2.9 Angstrom Resolution Cryo-EM 3-D Reconstruction of Close-packed PCV2 Virus-like Particles | |||||||||
 Map data Map data | Reconstruction of porcine circovirus 2 (PCV2) virus-like particles | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | de novo initial model / consensus criterion / gold-standard FSC / true FSC / cross-validation | |||||||||
| Function / homology |  Function and homology information Function and homology informationviral capsid assembly / T=1 icosahedral viral capsid / viral penetration into host nucleus / host cell / endocytosis involved in viral entry into host cell / virion attachment to host cell / host cell nucleus Similarity search - Function | |||||||||
| Biological species |   Porcine circovirus 2 Porcine circovirus 2 | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.9 Å | |||||||||
 Authors Authors | Liu Z / Guo F / Wang F / Li TC / Jiang W | |||||||||
 Citation Citation |  Journal: Structure / Year: 2016 Journal: Structure / Year: 2016Title: 2.9 Å Resolution Cryo-EM 3D Reconstruction of Close-Packed Virus Particles. Authors: Zheng Liu / Fei Guo / Feng Wang / Tian-Cheng Li / Wen Jiang /   Abstract: Single-particle cryoelectron microscopy typically discards close-packed particle images as unusable data. Here, we report an image processing strategy and case study of obtaining near-atomic ...Single-particle cryoelectron microscopy typically discards close-packed particle images as unusable data. Here, we report an image processing strategy and case study of obtaining near-atomic resolution 3D reconstructions from close-packed particles. Multiple independent de novo initial models were constructed to determine and cross-validate the particle parameters. The particles with consistent views were further refined including not only Euler angles and center positions but also defocus, astigmatism, beam tilt, and overall and anisotropic magnification. We demonstrated this strategy with a 2.9 Å resolution reconstruction of a 1.67 MDa virus-like particle of a circovirus, PCV2, recorded on 86 photographic films. The map resolution was further validated with a phase-randomization test and local resolution assessment, and the atomic model was validated with MolProbity and EMRinger. Close-packed virus particles were thus shown not only to be useful for high-resolution 3D reconstructions but also to allow data collection at significantly improved throughput for near-atomic resolution reconstructions. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_6555.map.gz emd_6555.map.gz | 108 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-6555-v30.xml emd-6555-v30.xml emd-6555.xml emd-6555.xml | 12.5 KB 12.5 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_6555.png emd_6555.png | 546.1 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-6555 http://ftp.pdbj.org/pub/emdb/structures/EMD-6555 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6555 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6555 | HTTPS FTP |
-Validation report
| Summary document |  emd_6555_validation.pdf.gz emd_6555_validation.pdf.gz | 367.6 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_6555_full_validation.pdf.gz emd_6555_full_validation.pdf.gz | 367.2 KB | Display | |
| Data in XML |  emd_6555_validation.xml.gz emd_6555_validation.xml.gz | 7 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-6555 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-6555 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-6555 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-6555 | HTTPS FTP |
-Related structure data
| Related structure data |  3jciMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_6555.map.gz / Format: CCP4 / Size: 122.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_6555.map.gz / Format: CCP4 / Size: 122.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
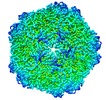
| Annotation | Reconstruction of porcine circovirus 2 (PCV2) virus-like particles | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.08 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
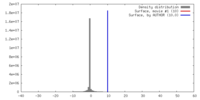
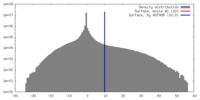
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Porcine circovirus PCV2 virus-like particles
| Entire | Name: Porcine circovirus PCV2 virus-like particles |
|---|---|
| Components |
|
-Supramolecule #1000: Porcine circovirus PCV2 virus-like particles
| Supramolecule | Name: Porcine circovirus PCV2 virus-like particles / type: sample / ID: 1000 / Number unique components: 1 |
|---|---|
| Molecular weight | Experimental: 1.67 MDa / Theoretical: 1.67 MDa |
-Supramolecule #1: Porcine circovirus 2
| Supramolecule | Name: Porcine circovirus 2 / type: virus / ID: 1 / Name.synonym: PCV2 / NCBI-ID: 85708 / Sci species name: Porcine circovirus 2 / Sci species strain: Yamagata / Virus type: VIRUS-LIKE PARTICLE / Virus isolate: STRAIN / Virus enveloped: No / Virus empty: Yes / Syn species name: PCV2 |
|---|---|
| Host (natural) | Organism:  |
| Host system | Organism:  Trichoplusia ni (cabbage looper) / Recombinant cell: Tn5 / Recombinant plasmid: AcPCV2-ORF2 Trichoplusia ni (cabbage looper) / Recombinant cell: Tn5 / Recombinant plasmid: AcPCV2-ORF2 |
| Molecular weight | Experimental: 1.67 MDa / Theoretical: 1.67 MDa |
| Virus shell | Shell ID: 1 / Diameter: 190 Å / T number (triangulation number): 1 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 3 mg/mL |
|---|---|
| Buffer | pH: 7.4 / Details: 1x PBS |
| Grid | Details: 400-mesh holey carbon grids (1.2/1.3 C-flat, Protochips) |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 90 % / Chamber temperature: 85 K / Instrument: GATAN CRYOPLUNGE 3 / Method: Blot for 5 seconds before plunging. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Temperature | Min: 80 K / Max: 100 K / Average: 90 K |
| Alignment procedure | Legacy - Astigmatism: Objective lens astigmatism was corrected at 250,000 magnification using a quadrupole stigmator. |
| Date | Feb 22, 2011 |
| Image recording | Category: FILM / Film or detector model: KODAK SO-163 FILM / Digitization - Scanner: NIKON SUPER COOLSCAN 9000 / Digitization - Sampling interval: 6.35 µm / Number real images: 141 / Average electron dose: 25 e/Å2 / Od range: 1 / Bits/pixel: 16 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.2 µm / Nominal magnification: 59000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Details | The particles were selected using the e2boxer.py program in EMAN2. CTF parameters were determined using fitctf2.py in JSPR. |
|---|---|
| CTF correction | Details: Each particle |
| Final reconstruction | Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 2.9 Å / Resolution method: OTHER / Software - Name: JSPR, EMAN2 Details: For 3D reconstruction, whole datasets were divided into even and odd halves and the initial de novo models and subsequent iterative refinements were all independently performed for each half ...Details: For 3D reconstruction, whole datasets were divided into even and odd halves and the initial de novo models and subsequent iterative refinements were all independently performed for each half dataset. Particles were selected from scanned micrograph images using e2boxer.py in EMAN2. The TEM instrument contrast transfer function parameters were determined automatically using fitctf2.py in JSPR and were then visually validated using the EMAN ctfit program. The datasets were then divided into two subsets (even and odd) and processed completely independently, including both de novo initial models and refinements. The images were first binned 4x to obtain initial models and particle parameters assuming icosahedral symmetry. De novo initial models were built using the random model approach. Random subsets of particles were assigned random initial orientations and iteratively refined until convergence. Multi-model competitive refinements were used to choose the winning model (with most assigned particles) as corrective initial models for subsequent refinement. Particles with inconsistent/unstable view parameters in the initial refinements were excluded in further image processing. The orientation and center parameters were then transferred to the un-binned images for high-resolution refinements which included Simplex method-based orientation/center optimization and grid search-based refinement of defocus, astigmatism, beam tilt, and overall and anisotropic magnification of the images. All image refinement and reconstructions were performed with JSPR software that was built on EMAN2 and EMAN library functions and programs. Number images used: 50352 |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)