+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-6313 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | 2.5A structure of lysozyme solved by MicroED | |||||||||
 Map data Map data | Lysozyme electron diffraction phased by molecular replacement. PDB ID 3J6K. | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | lysozyme / microcrystal | |||||||||
| Function / homology |  Function and homology information Function and homology informationLactose synthesis / Antimicrobial peptides / Neutrophil degranulation / beta-N-acetylglucosaminidase activity / cell wall macromolecule catabolic process / lysozyme / lysozyme activity / defense response to Gram-negative bacterium / killing of cells of another organism / defense response to Gram-positive bacterium ...Lactose synthesis / Antimicrobial peptides / Neutrophil degranulation / beta-N-acetylglucosaminidase activity / cell wall macromolecule catabolic process / lysozyme / lysozyme activity / defense response to Gram-negative bacterium / killing of cells of another organism / defense response to Gram-positive bacterium / defense response to bacterium / endoplasmic reticulum / extracellular space / identical protein binding / cytoplasm Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | electron crystallography / cryo EM / Resolution: 2.5 Å | |||||||||
 Authors Authors | Nannenga BL / Shi D / Leslie AGW / Gonen T | |||||||||
 Citation Citation |  Journal: Nat Methods / Year: 2014 Journal: Nat Methods / Year: 2014Title: High-resolution structure determination by continuous-rotation data collection in MicroED. Authors: Brent L Nannenga / Dan Shi / Andrew G W Leslie / Tamir Gonen /   Abstract: MicroED uses very small three-dimensional protein crystals and electron diffraction for structure determination. We present an improved data collection protocol for MicroED called 'continuous ...MicroED uses very small three-dimensional protein crystals and electron diffraction for structure determination. We present an improved data collection protocol for MicroED called 'continuous rotation'. Microcrystals are continuously rotated during data collection, yielding more accurate data. The method enables data processing with the crystallographic software tool MOSFLM, which resulted in improved resolution for the model protein lysozyme. These improvements are paving the way for the broad implementation and application of MicroED in structural biology. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_6313.map.gz emd_6313.map.gz | 3.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-6313-v30.xml emd-6313-v30.xml emd-6313.xml emd-6313.xml | 12 KB 12 KB | Display Display |  EMDB header EMDB header |
| Images |  400_6313.gif 400_6313.gif 80_6313.gif 80_6313.gif | 59.8 KB 4.2 KB | ||
| Masks |  emd_6313_msk_1.map emd_6313_msk_1.map | 965.4 KB |  Mask map Mask map | |
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-6313 http://ftp.pdbj.org/pub/emdb/structures/EMD-6313 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6313 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6313 | HTTPS FTP |
-Validation report
| Summary document |  emd_6313_validation.pdf.gz emd_6313_validation.pdf.gz | 412.7 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_6313_full_validation.pdf.gz emd_6313_full_validation.pdf.gz | 412.2 KB | Display | |
| Data in XML |  emd_6313_validation.xml.gz emd_6313_validation.xml.gz | 4.5 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-6313 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-6313 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-6313 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-6313 | HTTPS FTP |
-Related structure data
| Related structure data |  3j6kMC  6342C  5a3eC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_6313.map.gz / Format: CCP4 / Size: 3.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_6313.map.gz / Format: CCP4 / Size: 3.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Lysozyme electron diffraction phased by molecular replacement. PDB ID 3J6K. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. generated in cubic-lattice coordinate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X: 0.60768 Å / Y: 0.60768 Å / Z: 0.62033 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
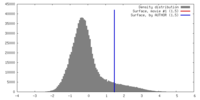
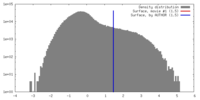
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 96 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Segmentation: This mask represents the map masked around the...
| Annotation | This mask represents the map masked around the asymmetric unit (PDB 3J6K) | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| File |  emd_6313_msk_1.map emd_6313_msk_1.map | ||||||||||||
| Projections & Slices |
| ||||||||||||
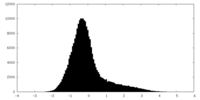
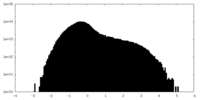
| Density Histograms |
- Sample components
Sample components
-Entire : Hen egg white lysozyme
| Entire | Name: Hen egg white lysozyme |
|---|---|
| Components |
|
-Supramolecule #1000: Hen egg white lysozyme
| Supramolecule | Name: Hen egg white lysozyme / type: sample / ID: 1000 / Details: lysozyme microcrystals / Oligomeric state: 1 / Number unique components: 1 |
|---|---|
| Molecular weight | Theoretical: 14.3 KDa |
-Macromolecule #1: Lysozyme C
| Macromolecule | Name: Lysozyme C / type: protein_or_peptide / ID: 1 / Name.synonym: lysozyme / Details: lysozyme microcrystals / Recombinant expression: No / Database: NCBI |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 14.3 KDa |
| Sequence | UniProtKB: Lysozyme C |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | electron crystallography |
| Aggregation state | 3D array |
- Sample preparation
Sample preparation
| Concentration | 200 mg/mL |
|---|---|
| Buffer | pH: 4.5 Details: 50 mM sodium acetate, 3.5 M sodium chloride, 15% PEG5000 |
| Grid | Details: 300 mesh copper grid with holey carbon support |
| Vitrification | Cryogen name: ETHANE / Chamber temperature: 100 K / Instrument: FEI VITROBOT MARK IV Method: Add crystals to grid for 30 seconds and blot for 10-12 seconds. |
| Details | batch crystallization |
| Crystal formation | Details: batch crystallization |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI F20 |
|---|---|
| Temperature | Min: 90 K / Max: 110 K / Average: 100 K |
| Date | Feb 4, 2014 |
| Image recording | Category: CCD / Film or detector model: TVIPS TEMCAM-F416 (4k x 4k) / Average electron dose: 0.1 e/Å2 / Camera length: 1500 Details: Raw diffraction images are available upon request. Contact the Gonen lab for access. |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: DIFFRACTION |
| Sample stage | Specimen holder model: GATAN LIQUID NITROGEN / Tilt angle min: -45 / Tilt angle max: 45 / Tilt series - Axis1 - Min angle: -45 ° / Tilt series - Axis1 - Max angle: 45 ° |
| Experimental equipment |  Model: Tecnai F20 / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 2.5 Å / Resolution method: DIFFRACTION PATTERN/LAYERLINES |
|---|---|
| Crystal parameters | Unit cell - A: 75.96 Å / Unit cell - B: 75.96 Å / Unit cell - C: 37.22 Å / Unit cell - γ: 90 ° / Unit cell - α: 90 ° / Unit cell - β: 90 ° / Space group: P 43 21 2 |
 Movie
Movie Controller
Controller









 X (Sec.)
X (Sec.) Y (Row.)
Y (Row.) Z (Col.)
Z (Col.)