[English] 日本語
 Yorodumi
Yorodumi- PDB-5y6p: Structure of the phycobilisome from the red alga Griffithsia pacifica -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 5y6p | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of the phycobilisome from the red alga Griffithsia pacifica | |||||||||
 Components Components |
| |||||||||
 Keywords Keywords | PHOTOSYNTHESIS / phycobilisome | |||||||||
| Function / homology | PHYCOCYANOBILIN / PHYCOERYTHROBILIN / PHYCOUROBILIN / PHYCOVIOLOBILIN Function and homology information Function and homology information | |||||||||
| Biological species |  Griffithsia pacifica (eukaryote) Griffithsia pacifica (eukaryote) | |||||||||
| Method | ELECTRON MICROSCOPY / single particle reconstruction / cryo EM / Resolution: 3.5 Å | |||||||||
 Authors Authors | Zhang, J. / Ma, J.F. / Liu, D.S. / Sun, S. / Sui, S.F. | |||||||||
 Citation Citation |  Journal: Nature / Year: 2017 Journal: Nature / Year: 2017Title: Structure of phycobilisome from the red alga Griffithsia pacifica. Authors: Jun Zhang / Jianfei Ma / Desheng Liu / Song Qin / Shan Sun / Jindong Zhao / Sen-Fang Sui /  Abstract: Life on Earth depends on photosynthesis for its conversion of solar energy to chemical energy. Photosynthetic organisms have developed a variety of light-harvesting systems to capture sunlight. The ...Life on Earth depends on photosynthesis for its conversion of solar energy to chemical energy. Photosynthetic organisms have developed a variety of light-harvesting systems to capture sunlight. The largest light-harvesting complex is the phycobilisome (PBS), the main light-harvesting antenna in cyanobacteria and red algae. It is composed of phycobiliproteins and linker proteins but the assembly mechanisms and energy transfer pathways of the PBS are not well understood. Here we report the structure of a 16.8-megadalton PBS from a red alga at 3.5 Å resolution obtained by single-particle cryo-electron microscopy. We modelled 862 protein subunits, including 4 linkers in the core, 16 rod-core linkers and 52 rod linkers, and located a total of 2,048 chromophores. This structure reveals the mechanisms underlying specific interactions between linkers and phycobiliproteins, and the formation of linker skeletons. These results provide a firm structural basis for our understanding of complex assembly and the mechanisms of energy transfer within the PBS. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  5y6p.cif.gz 5y6p.cif.gz | 28.7 MB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb5y6p.ent.gz pdb5y6p.ent.gz | Display |  PDB format PDB format | |
| PDBx/mmJSON format |  5y6p.json.gz 5y6p.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  5y6p_validation.pdf.gz 5y6p_validation.pdf.gz | 191.1 MB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  5y6p_full_validation.pdf.gz 5y6p_full_validation.pdf.gz | 202.2 MB | Display | |
| Data in XML |  5y6p_validation.xml.gz 5y6p_validation.xml.gz | 5.7 MB | Display | |
| Data in CIF |  5y6p_validation.cif.gz 5y6p_validation.cif.gz | 7.1 MB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/y6/5y6p https://data.pdbj.org/pub/pdb/validation_reports/y6/5y6p ftp://data.pdbj.org/pub/pdb/validation_reports/y6/5y6p ftp://data.pdbj.org/pub/pdb/validation_reports/y6/5y6p | HTTPS FTP |
-Related structure data
| Related structure data |  6769MC  6758C  6759C  6760C  6761C  6762C  6763C  6764C  6765C  6766C  6767C  6768C M: map data used to model this data C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
|
|---|---|
| 1 |
|
- Components
Components
+Protein , 23 types, 190 molecules A1B1C1D1E1F1G1I1K1N1P1R1T1X1Z1b1d1f1i1k1o1q1s1u1w1y1H1J1L1M1...
-R-phycoerythrin ... , 2 types, 672 molecules e2g2i2k2m2o2q2s2u2w2y2E2G2I2K2M2O2Q2S2U2W2Y2A3C3E3G3I3K3M3O3...
| #10: Protein | Mass: 17763.820 Da / Num. of mol.: 326 / Source method: isolated from a natural source / Source: (natural)  Griffithsia pacifica (eukaryote) Griffithsia pacifica (eukaryote)#11: Protein | Mass: 18496.031 Da / Num. of mol.: 346 / Source method: isolated from a natural source / Source: (natural)  Griffithsia pacifica (eukaryote) Griffithsia pacifica (eukaryote) |
|---|
-Non-polymers , 4 types, 2048 molecules 

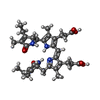
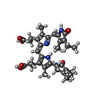



| #26: Chemical | ChemComp-PEB / #27: Chemical | ChemComp-CYC / #28: Chemical | ChemComp-PUB / #29: Chemical | ChemComp-PVN / |
|---|
-Details
| Has protein modification | Y |
|---|
-Experimental details
-Experiment
| Experiment | Method: ELECTRON MICROSCOPY |
|---|---|
| EM experiment | Aggregation state: PARTICLE / 3D reconstruction method: single particle reconstruction |
- Sample preparation
Sample preparation
| Component | Name: phycobilisome / Type: COMPLEX / Entity ID: #1-#25 / Source: NATURAL |
|---|---|
| Molecular weight | Value: 16.8 MDa / Experimental value: NO |
| Source (natural) | Organism:  Griffithsia pacifica (eukaryote) Griffithsia pacifica (eukaryote) |
| Buffer solution | pH: 7 |
| Specimen | Conc.: 1 mg/ml / Embedding applied: NO / Shadowing applied: NO / Staining applied: NO / Vitrification applied: YES |
| Vitrification | Cryogen name: ETHANE / Humidity: 100 % / Chamber temperature: 289.15 K |
- Electron microscopy imaging
Electron microscopy imaging
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
|---|---|
| Microscopy | Model: FEI TITAN KRIOS |
| Electron gun | Electron source:  FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM |
| Electron lens | Mode: BRIGHT FIELD |
| Image recording | Electron dose: 50 e/Å2 / Film or detector model: GATAN K2 SUMMIT (4k x 4k) |
- Processing
Processing
| CTF correction | Type: NONE |
|---|---|
| 3D reconstruction | Resolution: 3.5 Å / Resolution method: FSC 0.143 CUT-OFF / Num. of particles: 53403 / Symmetry type: POINT |
 Movie
Movie Controller
Controller






 PDBj
PDBj



