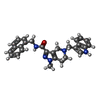
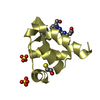
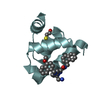
Entry Database : PDB / ID : 5l87Title Targeting the PEX14-PEX5 interaction by small molecules provides novel therapeutic routes to treat trypanosomiases. Peroxin 14 Keywords / / / / / Function / homology Function Domain/homology Component
/ / / / / / / / / / / / / Biological species Trypanosoma brucei brucei (eukaryote)Method / / / Resolution : 0.87 Å Authors Dawidowski, M. / Emmanouilidis, L. / Sattler, M. / Popowicz, G.M. Funding support Organization Grant number Country German Research Foundation FOR1905 ER178/4-1
Journal : Science / Year : 2017Title : Inhibitors of PEX14 disrupt protein import into glycosomes and kill Trypanosoma parasites.Authors: Dawidowski, M. / Emmanouilidis, L. / Kalel, V.C. / Tripsianes, K. / Schorpp, K. / Hadian, K. / Kaiser, M. / Maser, P. / Kolonko, M. / Tanghe, S. / Rodriguez, A. / Schliebs, W. / Erdmann, R. ... Authors : Dawidowski, M. / Emmanouilidis, L. / Kalel, V.C. / Tripsianes, K. / Schorpp, K. / Hadian, K. / Kaiser, M. / Maser, P. / Kolonko, M. / Tanghe, S. / Rodriguez, A. / Schliebs, W. / Erdmann, R. / Sattler, M. / Popowicz, G.M. History Deposition Jun 7, 2016 Deposition site / Processing site Revision 1.0 Mar 8, 2017 Provider / Type Revision 1.1 Apr 12, 2017 Group Revision 1.2 Oct 16, 2019 Group / Data collection / Category / reflns_shell / Item Revision 1.3 Jan 10, 2024 Group / Database references / Refinement descriptionCategory chem_comp_atom / chem_comp_bond ... chem_comp_atom / chem_comp_bond / database_2 / pdbx_initial_refinement_model Item / _database_2.pdbx_database_accession
Show all Show less
 Yorodumi
Yorodumi Open data
Open data Basic information
Basic information Components
Components Keywords
Keywords Function and homology information
Function and homology information
 X-RAY DIFFRACTION /
X-RAY DIFFRACTION /  SYNCHROTRON /
SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 0.87 Å
MOLECULAR REPLACEMENT / Resolution: 0.87 Å  Authors
Authors Germany, 1items
Germany, 1items  Citation
Citation Journal: Science / Year: 2017
Journal: Science / Year: 2017 Structure visualization
Structure visualization Molmil
Molmil Jmol/JSmol
Jmol/JSmol Downloads & links
Downloads & links Download
Download 5l87.cif.gz
5l87.cif.gz PDBx/mmCIF format
PDBx/mmCIF format pdb5l87.ent.gz
pdb5l87.ent.gz PDB format
PDB format 5l87.json.gz
5l87.json.gz PDBx/mmJSON format
PDBx/mmJSON format Other downloads
Other downloads 5l87_validation.pdf.gz
5l87_validation.pdf.gz wwPDB validaton report
wwPDB validaton report 5l87_full_validation.pdf.gz
5l87_full_validation.pdf.gz 5l87_validation.xml.gz
5l87_validation.xml.gz 5l87_validation.cif.gz
5l87_validation.cif.gz https://data.pdbj.org/pub/pdb/validation_reports/l8/5l87
https://data.pdbj.org/pub/pdb/validation_reports/l8/5l87 ftp://data.pdbj.org/pub/pdb/validation_reports/l8/5l87
ftp://data.pdbj.org/pub/pdb/validation_reports/l8/5l87



 Links
Links Assembly
Assembly
 Components
Components

 X-RAY DIFFRACTION / Number of used crystals: 1
X-RAY DIFFRACTION / Number of used crystals: 1  Sample preparation
Sample preparation SYNCHROTRON / Site:
SYNCHROTRON / Site:  ESRF
ESRF  / Beamline: ID29 / Wavelength: 0.8726 Å
/ Beamline: ID29 / Wavelength: 0.8726 Å Processing
Processing MOLECULAR REPLACEMENT
MOLECULAR REPLACEMENT Movie
Movie Controller
Controller








 PDBj
PDBj