[English] 日本語
 Yorodumi
Yorodumi- PDB-5k9c: Crystal structure of human dihydroorotate dehydrogenase with ML390 -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 5k9c | ||||||
|---|---|---|---|---|---|---|---|
| Title | Crystal structure of human dihydroorotate dehydrogenase with ML390 | ||||||
 Components Components | Dihydroorotate dehydrogenase (quinone), mitochondrial | ||||||
 Keywords Keywords | OXIDOREDUCTASE/OXIDOREDUCTASE INHIBITOR / Oxidoreductase / alpha/beta barrel / inhibitor / OXIDOREDUCTASE-OXIDOREDUCTASE INHIBITOR complex | ||||||
| Function / homology |  Function and homology information Function and homology informationpyrimidine ribonucleotide biosynthetic process / dihydroorotate dehydrogenase (quinone) / dihydroorotate dehydrogenase (quinone) activity / dihydroorotate dehydrogenase activity / dihydroorotase activity / Pyrimidine biosynthesis / UDP biosynthetic process / 'de novo' UMP biosynthetic process / 'de novo' pyrimidine nucleobase biosynthetic process / mitochondrial inner membrane ...pyrimidine ribonucleotide biosynthetic process / dihydroorotate dehydrogenase (quinone) / dihydroorotate dehydrogenase (quinone) activity / dihydroorotate dehydrogenase activity / dihydroorotase activity / Pyrimidine biosynthesis / UDP biosynthetic process / 'de novo' UMP biosynthetic process / 'de novo' pyrimidine nucleobase biosynthetic process / mitochondrial inner membrane / mitochondrion / nucleoplasm / cytosol Similarity search - Function | ||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / MOLECULAR REPLACEMENT /  molecular replacement / Resolution: 1.66 Å molecular replacement / Resolution: 1.66 Å | ||||||
 Authors Authors | Lewis, T.A. / Sykes, D.B. / Law, J.M. / Munoz, B. / Scadden, D.T. / Rustiguel, J.K. / Nonato, M.C. / Schreiber, S.L. | ||||||
 Citation Citation |  Journal: ACS Med Chem Lett / Year: 2016 Journal: ACS Med Chem Lett / Year: 2016Title: Development of ML390: A Human DHODH Inhibitor That Induces Differentiation in Acute Myeloid Leukemia. Authors: Lewis, T.A. / Sykes, D.B. / Law, J.M. / Munoz, B. / Rustiguel, J.K. / Nonato, M.C. / Scadden, D.T. / Schreiber, S.L. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  5k9c.cif.gz 5k9c.cif.gz | 231.3 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb5k9c.ent.gz pdb5k9c.ent.gz | 187.1 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  5k9c.json.gz 5k9c.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/k9/5k9c https://data.pdbj.org/pub/pdb/validation_reports/k9/5k9c ftp://data.pdbj.org/pub/pdb/validation_reports/k9/5k9c ftp://data.pdbj.org/pub/pdb/validation_reports/k9/5k9c | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  5k9dC  1d3gS C: citing same article ( S: Starting model for refinement |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
| ||||||||
| Components on special symmetry positions |
|
- Components
Components
-Protein , 1 types, 1 molecules A
| #1: Protein | Mass: 39943.613 Da / Num. of mol.: 1 / Fragment: residues 29-395 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: DHODH / Plasmid: pET28a / Details (production host): pET28a modified vector / Production host: Homo sapiens (human) / Gene: DHODH / Plasmid: pET28a / Details (production host): pET28a modified vector / Production host:  References: UniProt: Q02127, dihydroorotate dehydrogenase (quinone) |
|---|
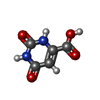
-Non-polymers , 8 types, 351 molecules 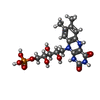













| #2: Chemical | ChemComp-FNR / | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| #3: Chemical | ChemComp-ORO / | ||||||||
| #4: Chemical | ChemComp-MLJ / ~{ | ||||||||
| #5: Chemical | | #6: Chemical | #7: Chemical | #8: Chemical | ChemComp-GOL / | #9: Water | ChemComp-HOH / | |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 3.43 Å3/Da / Density % sol: 64.2 % |
|---|---|
| Crystal grow | Temperature: 298 K / Method: vapor diffusion, sitting drop / pH: 4.8 Details: 0.1 M sodium acetate trihydrate pH 4.8 1.9 M ammonium sulfate 30% (v/v) glycerol PH range: 4.8 |
-Data collection
| Diffraction | Mean temperature: 100 K | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  LNLS LNLS  / Beamline: W01B-MX2 / Wavelength: 1.4586 Å / Beamline: W01B-MX2 / Wavelength: 1.4586 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Detector | Type: DECTRIS PILATUS3 2M / Detector: PIXEL / Date: Mar 10, 2015 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Radiation | Monochromator: Si(111) double-crystal monochromator and toroidal bendable mirror Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Radiation wavelength | Wavelength: 1.4586 Å / Relative weight: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reflection | Resolution: 1.66→25.952 Å / Num. obs: 67579 / % possible obs: 99.8 % / Redundancy: 15.1 % / Biso Wilson estimate: 14.33 Å2 / CC1/2: 0.998 / Rsym value: 0.097 / Net I/σ(I): 18.7 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reflection shell |
|
-Phasing
| Phasing | Method:  molecular replacement molecular replacement | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Phasing MR |
|
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: 1D3G Resolution: 1.66→25.952 Å / SU ML: 0.1 / Cross valid method: FREE R-VALUE / σ(F): 1.33 / Phase error: 12.65 Details: THE AUTHORS STATE THAT ELECTRON DENSITY ANALYSIS REVEALED THAT THE MAIN CHAIN HAD A DOUBLE CONFORMATION ( FROM RESIDUE 211 TO 226), DUE TO ITS EXPECTED FLEXIBILITY (AS A CATALYTIC LOOP). ...Details: THE AUTHORS STATE THAT ELECTRON DENSITY ANALYSIS REVEALED THAT THE MAIN CHAIN HAD A DOUBLE CONFORMATION ( FROM RESIDUE 211 TO 226), DUE TO ITS EXPECTED FLEXIBILITY (AS A CATALYTIC LOOP). VAL213 SHOWED A VERY CLEAR ELECTRON DENSITY IN CONFORMATION B, BUT IN CONTRAST, CONFORMATION A WAS DOMINANT FOR RESIDUES FROM 214 TO 218. THE VAL213 CONFORMER AND THE SER214 A CONFORMER WERE CONFLICTING AS A CONSEQUENCE OF THE DIFFERENT LOOP ARRANGEMENTS (OPEN AND CLOSED CONFORMATIONS). TAKING ALL THESE INTO ACCOUNT, THE AUTHORS STATE THAT THE VAL213 DOUBLE CONFORMATION IS THE SOLUTION THAT BETTER FITS THE DATASET AND THEREFORE, VAL213 (B CONFORMER) AND SER214 (A CONFORMER) ARE NOT LINKED.
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Shrinkage radii: 0.9 Å / VDW probe radii: 1.11 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso max: 136.79 Å2 / Biso mean: 23.5645 Å2 / Biso min: 5.99 Å2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: final / Resolution: 1.66→25.952 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Refine-ID: X-RAY DIFFRACTION / Total num. of bins used: 23
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS params. | Method: refined / Refine-ID: X-RAY DIFFRACTION
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS group |
|
 Movie
Movie Controller
Controller












 PDBj
PDBj







