[English] 日本語
 Yorodumi
Yorodumi- EMDB-5532: Single particle tomography of TRiC chaperonin with internalized s... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-5532 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Single particle tomography of TRiC chaperonin with internalized substrate | |||||||||
 Map data Map data | Single Particle Tomography average of TRiC chaperonin incubated with mhttQ51 | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Mutant huntingtin / TRiC chaperonin / Single Particle Tomography / cryo electron microscopy / amyloid | |||||||||
| Biological species |  | |||||||||
| Method | subtomogram averaging / cryo EM / Resolution: 50.0 Å | |||||||||
 Authors Authors | Shahmoradian SH / Galaz JG / Schmid MF / Cong Y / Ma B / Spiess C / Frydman J / Ludtke SJ / Chiu W | |||||||||
 Citation Citation |  Journal: Elife / Year: 2013 Journal: Elife / Year: 2013Title: TRiC's tricks inhibit huntingtin aggregation. Authors: Sarah H Shahmoradian / Jesus G Galaz-Montoya / Michael F Schmid / Yao Cong / Boxue Ma / Christoph Spiess / Judith Frydman / Steven J Ludtke / Wah Chiu /  Abstract: In Huntington's disease, a mutated version of the huntingtin protein leads to cell death. Mutant huntingtin is known to aggregate, a process that can be inhibited by the eukaryotic chaperonin TRiC ...In Huntington's disease, a mutated version of the huntingtin protein leads to cell death. Mutant huntingtin is known to aggregate, a process that can be inhibited by the eukaryotic chaperonin TRiC (TCP1-ring complex) in vitro and in vivo. A structural understanding of the genesis of aggregates and their modulation by cellular chaperones could facilitate the development of therapies but has been hindered by the heterogeneity of amyloid aggregates. Using cryo-electron microscopy (cryoEM) and single particle cryo-electron tomography (SPT) we characterize the growth of fibrillar aggregates of mutant huntingtin exon 1 containing an expanded polyglutamine tract with 51 residues (mhttQ51), and resolve 3-D structures of the chaperonin TRiC interacting with mhttQ51. We find that TRiC caps mhttQ51 fibril tips via the apical domains of its subunits, and also encapsulates smaller mhtt oligomers within its chamber. These two complementary mechanisms provide a structural description for TRiC's inhibition of mhttQ51 aggregation in vitro. DOI:http://dx.doi.org/10.7554/eLife.00710.001. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_5532.map.gz emd_5532.map.gz | 3.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-5532-v30.xml emd-5532-v30.xml emd-5532.xml emd-5532.xml | 9.9 KB 9.9 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_5532.png emd_5532.png emd_5532_1.png emd_5532_1.png | 148.1 KB 126.7 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-5532 http://ftp.pdbj.org/pub/emdb/structures/EMD-5532 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-5532 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-5532 | HTTPS FTP |
-Validation report
| Summary document |  emd_5532_validation.pdf.gz emd_5532_validation.pdf.gz | 78.4 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_5532_full_validation.pdf.gz emd_5532_full_validation.pdf.gz | 77.5 KB | Display | |
| Data in XML |  emd_5532_validation.xml.gz emd_5532_validation.xml.gz | 492 B | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-5532 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-5532 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-5532 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-5532 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_5532.map.gz / Format: CCP4 / Size: 3.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_5532.map.gz / Format: CCP4 / Size: 3.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Single Particle Tomography average of TRiC chaperonin incubated with mhttQ51 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 4.401 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
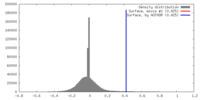
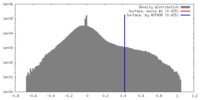
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : TRiC chaperonin + mhttQ51
| Entire | Name: TRiC chaperonin + mhttQ51 |
|---|---|
| Components |
|
-Supramolecule #1000: TRiC chaperonin + mhttQ51
| Supramolecule | Name: TRiC chaperonin + mhttQ51 / type: sample / ID: 1000 / Number unique components: 2 |
|---|---|
| Molecular weight | Theoretical: 1.01 MDa |
-Macromolecule #1: TCP-1 Ring Complex with mutant huntingtin Q51 exon-1
| Macromolecule | Name: TCP-1 Ring Complex with mutant huntingtin Q51 exon-1 / type: protein_or_peptide / ID: 1 / Name.synonym: TRiC with mhtt / Recombinant expression: No / Database: NCBI |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 1.01 MDa |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Grid | Details: 200 mesh copper Quantifoil grid, glow discharged |
|---|---|
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Instrument: FEI VITROBOT MARK III |
- Electron microscopy
Electron microscopy
| Microscope | JEOL 2100 |
|---|---|
| Details | Serial EM software |
| Date | Mar 19, 2009 |
| Image recording | Category: CCD / Film or detector model: GENERIC CCD / Digitization - Sampling interval: 2 µm / Number real images: 121 / Average electron dose: 62 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source: LAB6 |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 5.0 µm / Nominal defocus min: 5.0 µm / Nominal magnification: 25000 |
| Sample stage | Specimen holder model: GATAN LIQUID NITROGEN / Tilt series - Axis1 - Min angle: -60 ° / Tilt series - Axis1 - Max angle: 60 ° |
- Image processing
Image processing
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 50.0 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: EMAN2 / Number subtomograms used: 7 |
|---|---|
| Final 3D classification | Number classes: 1 |
 Movie
Movie Controller
Controller


 UCSF Chimera
UCSF Chimera









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)