+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Compact CVB1-VLP (Tween80) | |||||||||||||||
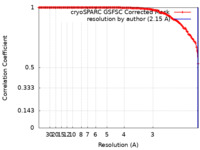
 Map data Map data | Density map of compact CVB1 VLP in presence of Tween80 resolved to 2.15 A resolution. | |||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | coxsackievirus B1 / vaccine / Tween80 / VIRUS LIKE PARTICLE | |||||||||||||||
| Function / homology |  Function and homology information Function and homology informationsymbiont-mediated suppression of host cytoplasmic pattern recognition receptor signaling pathway via inhibition of RIG-I activity / picornain 2A / symbiont-mediated suppression of host mRNA export from nucleus / symbiont genome entry into host cell via pore formation in plasma membrane / picornain 3C / T=pseudo3 icosahedral viral capsid / host cell cytoplasmic vesicle membrane / endocytosis involved in viral entry into host cell / nucleoside-triphosphate phosphatase / channel activity ...symbiont-mediated suppression of host cytoplasmic pattern recognition receptor signaling pathway via inhibition of RIG-I activity / picornain 2A / symbiont-mediated suppression of host mRNA export from nucleus / symbiont genome entry into host cell via pore formation in plasma membrane / picornain 3C / T=pseudo3 icosahedral viral capsid / host cell cytoplasmic vesicle membrane / endocytosis involved in viral entry into host cell / nucleoside-triphosphate phosphatase / channel activity / monoatomic ion transmembrane transport / DNA replication / RNA helicase activity / induction by virus of host autophagy / RNA-directed RNA polymerase / viral RNA genome replication / cysteine-type endopeptidase activity / RNA-dependent RNA polymerase activity / virus-mediated perturbation of host defense response / DNA-templated transcription / host cell nucleus / virion attachment to host cell / structural molecule activity / ATP hydrolysis activity / proteolysis / RNA binding / ATP binding / membrane / metal ion binding Similarity search - Function | |||||||||||||||
| Biological species |  Coxsackievirus B1 Coxsackievirus B1 | |||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.15 Å | |||||||||||||||
 Authors Authors | Plavec Z / Butcher SJ | |||||||||||||||
| Funding support |  Finland, 4 items Finland, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Res Sq / Year: 2024 Journal: Res Sq / Year: 2024Title: Comparison of structure and immunogenicity of CVB1-VLP and inactivated CVB1 vaccine candidates. Authors: Saana Soppela / Zlatka Plavec / Stina Gröhn / Minne Jartti / Sami Oikarinen / Mira Laajala / Varpu Marjomaki / Sarah J Butcher / Minna M Hankaniemi Abstract: Coxsackievirus B1 (CVB1) is a common cause of acute and chronic myocarditis, dilated cardiomyopathy and aseptic meningitis. However, no CVB-vaccines are available for human use. In this study, we ...Coxsackievirus B1 (CVB1) is a common cause of acute and chronic myocarditis, dilated cardiomyopathy and aseptic meningitis. However, no CVB-vaccines are available for human use. In this study, we investigated the immunogenicity of virus-like particle (VLP) and inactivated whole-virus vaccines for CVB1 when administrated to mice via either subcutaneous or intranasal routes formulated with and without commercial and experimental adjuvants. Here, the potential of utilizing epigallocatechin-3-gallate (EGCG) as a mucosal adjuvant synergistically with its ability to inactivate the virus were investigated. EGCG had promising adjuvant properties for CVB1-VLP when administered via the parenteral route but limited efficacy via intranasal administration. However, intranasal administration of the formalin-inactivated virus induced high CVB1-specific humoral, cellular, and mucosal immune responses. Also, based on CVB1-specific IgG-antibody responses, we conclude that CVB1-VLP can be taken up by immune cells when administrated intranasally and further structural engineering for the VLP may increase the mucosal immunogenicity. The preparations contained mixtures of compact and expanded A particles with 85% expanded in the formalin-inactivated virus, but only 52% in the VLP observed by cryogenic electron microscopy. To correlate the structure to immunogenicity, we solved the structures of the CVB1-VLP and the formalin-inactivated CVB1 virus at resolutions ranging from 2.15 A to 4.1 A for the expanded and compact VLP and virus particles by image reconstruction. These structures can be used in designing mutations increasing the stability and immunogenicity of CVB1-VLP in the future. Overall, our results highlight the potential of using formalin inactivated CVB1 vaccine in mucosal immunization programs and provide important information for future development of VLP-based vaccines against all enteroviruses. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_50497.map.gz emd_50497.map.gz | 112.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-50497-v30.xml emd-50497-v30.xml emd-50497.xml emd-50497.xml | 19.3 KB 19.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_50497_fsc.xml emd_50497_fsc.xml | 13 KB | Display |  FSC data file FSC data file |
| Images |  emd_50497.png emd_50497.png | 70.9 KB | ||
| Filedesc metadata |  emd-50497.cif.gz emd-50497.cif.gz | 6 KB | ||
| Others |  emd_50497_half_map_1.map.gz emd_50497_half_map_1.map.gz emd_50497_half_map_2.map.gz emd_50497_half_map_2.map.gz | 220.1 MB 220.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-50497 http://ftp.pdbj.org/pub/emdb/structures/EMD-50497 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-50497 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-50497 | HTTPS FTP |
-Validation report
| Summary document |  emd_50497_validation.pdf.gz emd_50497_validation.pdf.gz | 1 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_50497_full_validation.pdf.gz emd_50497_full_validation.pdf.gz | 1 MB | Display | |
| Data in XML |  emd_50497_validation.xml.gz emd_50497_validation.xml.gz | 22 KB | Display | |
| Data in CIF |  emd_50497_validation.cif.gz emd_50497_validation.cif.gz | 28.6 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-50497 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-50497 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-50497 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-50497 | HTTPS FTP |
-Related structure data
| Related structure data |  9fjcMC  9fjdC  9fjeC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_50497.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_50497.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Density map of compact CVB1 VLP in presence of Tween80 resolved to 2.15 A resolution. | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.06 Å | ||||||||||||||||||||||||||||||||||||
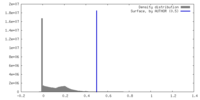
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Half map of compact CVB1 VLP in presence of Tween80.
| File | emd_50497_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map of compact CVB1 VLP in presence of Tween80. | ||||||||||||
| Projections & Slices |
| ||||||||||||
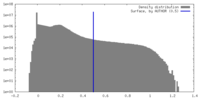
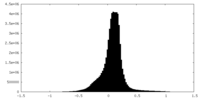
| Density Histograms |
-Half map: Half map of compact CVB1 VLP in presence of Tween80.
| File | emd_50497_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map of compact CVB1 VLP in presence of Tween80. | ||||||||||||
| Projections & Slices |
| ||||||||||||
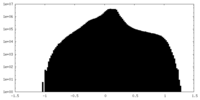
| Density Histograms |
- Sample components
Sample components
-Entire : Coxsackievirus B1
| Entire | Name:  Coxsackievirus B1 Coxsackievirus B1 |
|---|---|
| Components |
|
-Supramolecule #1: Coxsackievirus B1
| Supramolecule | Name: Coxsackievirus B1 / type: virus / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 Details: Coxsackievirus B1 virus-like particle based on CVB1-10796, isolated from Argentina. NCBI-ID: 12071 / Sci species name: Coxsackievirus B1 / Virus type: VIRUS-LIKE PARTICLE / Virus isolate: OTHER / Virus enveloped: No / Virus empty: Yes |
|---|---|
| Host (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Virus shell | Shell ID: 1 / Name: capsid / Diameter: 30.0 Å |
-Macromolecule #1: Capsid protein VP1
| Macromolecule | Name: Capsid protein VP1 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Coxsackievirus B1 Coxsackievirus B1 |
| Molecular weight | Theoretical: 25.415807 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: YHSRSESSIE NFLCRSACVY YATYTNNSKK GFAEWVINTR QVAQLRRKLE LFTYLRFDLE LTFVITSAQQ PSTASSVDAP VQTHQIMYV PPGGPVPTKV KDYAWQTSTN PSVFWTEGNA PPRMSIPFIS IGNAYSCFYD GWTQFSRNGV YGINTLNNMG T LYMRHVNE ...String: YHSRSESSIE NFLCRSACVY YATYTNNSKK GFAEWVINTR QVAQLRRKLE LFTYLRFDLE LTFVITSAQQ PSTASSVDAP VQTHQIMYV PPGGPVPTKV KDYAWQTSTN PSVFWTEGNA PPRMSIPFIS IGNAYSCFYD GWTQFSRNGV YGINTLNNMG T LYMRHVNE AGQGPIKSTV RIYFKPKHVK AWVPRPPRLC QYEKQKNVNF SPIGVTTSRT DIITT UniProtKB: Genome polyprotein |
-Macromolecule #2: Capsid protein VP2
| Macromolecule | Name: Capsid protein VP2 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Coxsackievirus B1 Coxsackievirus B1 |
| Molecular weight | Theoretical: 27.839439 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: VRSITLGNST ITTQECANVV VGYGVWPEYL KDNEATAEDQ PTQPDVATCR FYTLESVQWM KNSAGWWWKL PDALSQMGLF GQNMQYHYL GRTGYTIHVQ CNASKFHQGC LLVVCVPEAE MGCSNLNNTP EFSELSGGDS ARMFTDTQVG ESNAKKVQTA V WNAGMGVG ...String: VRSITLGNST ITTQECANVV VGYGVWPEYL KDNEATAEDQ PTQPDVATCR FYTLESVQWM KNSAGWWWKL PDALSQMGLF GQNMQYHYL GRTGYTIHVQ CNASKFHQGC LLVVCVPEAE MGCSNLNNTP EFSELSGGDS ARMFTDTQVG ESNAKKVQTA V WNAGMGVG VGNLTIFPHQ WINLRTNNSA TLVMPYINSV PMDNMFRHNN LTLMIIPFVP LNYSEGSSPY VPITVTIAPM CA EYNGLRL ASNQ UniProtKB: Genome polyprotein |
-Macromolecule #3: Capsid protein VP3
| Macromolecule | Name: Capsid protein VP3 / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Coxsackievirus B1 Coxsackievirus B1 |
| Molecular weight | Theoretical: 26.328764 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: GLPVMTTPGS TQFLTSDDFQ SPSAMPQFDV TPEMQIPGRV NNLMEIAEVD SVVPVNNTED NVSSLKAYQI PVQSNSDNGK QVFGFPLQP GANNVLNRTL LGEILNYYTH WSGSIKLTFM FCGSAMATGK FLLAYSPPGA GVPKNRKDAM LGTHVIWDVG L QSSCVLCV ...String: GLPVMTTPGS TQFLTSDDFQ SPSAMPQFDV TPEMQIPGRV NNLMEIAEVD SVVPVNNTED NVSSLKAYQI PVQSNSDNGK QVFGFPLQP GANNVLNRTL LGEILNYYTH WSGSIKLTFM FCGSAMATGK FLLAYSPPGA GVPKNRKDAM LGTHVIWDVG L QSSCVLCV PWISQTHYRY VVEDEYTAAG YVTCWYQTNI VVPADVQSSC DILCFVSACN DFSVRMLKDT PFIRQDTFYQ UniProtKB: Genome polyprotein |
-Macromolecule #4: Capsid protein VP4
| Macromolecule | Name: Capsid protein VP4 / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Coxsackievirus B1 Coxsackievirus B1 |
| Molecular weight | Theoretical: 1.96706 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: YTNINYYKDA ASNSANR UniProtKB: Genome polyprotein |
-Macromolecule #5: PALMITIC ACID
| Macromolecule | Name: PALMITIC ACID / type: ligand / ID: 5 / Number of copies: 1 / Formula: PLM |
|---|---|
| Molecular weight | Theoretical: 256.424 Da |
| Chemical component information |  ChemComp-PLM: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: CONTINUOUS / Support film - Film thickness: 200 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 10 sec. |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 85 % / Chamber temperature: 295 K / Instrument: LEICA EM GP |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 63.368 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.8000000000000003 µm / Nominal defocus min: 0.1 µm / Nominal magnification: 105000 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)