[English] 日本語
 Yorodumi
Yorodumi- PDB-4mr8: Crystal structure of the extracellular domain of human GABA(B) re... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 4mr8 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Crystal structure of the extracellular domain of human GABA(B) receptor bound to the antagonist CGP35348 | |||||||||
 Components Components | (Gamma-aminobutyric acid type B receptor subunit ...) x 2 | |||||||||
 Keywords Keywords | SIGNALING PROTEIN/ANTAGONIST / heterodimeric protein complex / Venus Flytrap module / neurotransmitter receptor / SIGNALING PROTEIN-ANTAGONIST complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationnegative regulation of gamma-aminobutyric acid secretion / GABA B receptor activation / G protein-coupled GABA receptor complex / : / neuron-glial cell signaling / : / G protein-coupled receptor heterodimeric complex / negative regulation of dopamine secretion / G protein-coupled GABA receptor activity / negative regulation of epinephrine secretion ...negative regulation of gamma-aminobutyric acid secretion / GABA B receptor activation / G protein-coupled GABA receptor complex / : / neuron-glial cell signaling / : / G protein-coupled receptor heterodimeric complex / negative regulation of dopamine secretion / G protein-coupled GABA receptor activity / negative regulation of epinephrine secretion / positive regulation of growth hormone secretion / extracellular matrix protein binding / GABA receptor complex / negative regulation of adenylate cyclase activity / Class C/3 (Metabotropic glutamate/pheromone receptors) / negative regulation of synaptic transmission / positive regulation of glutamate secretion / gamma-aminobutyric acid signaling pathway / synaptic transmission, GABAergic / axolemma / dendritic shaft / response to nicotine / adenylate cyclase-inhibiting G protein-coupled receptor signaling pathway / GABA-ergic synapse / mitochondrial membrane / Schaffer collateral - CA1 synapse / Activation of G protein gated Potassium channels / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits / synaptic vesicle / osteoblast differentiation / transmembrane signaling receptor activity / presynaptic membrane / response to ethanol / G alpha (i) signalling events / dendritic spine / chemical synaptic transmission / postsynaptic membrane / neuron projection / G protein-coupled receptor signaling pathway / protein heterodimerization activity / negative regulation of cell population proliferation / neuronal cell body / endoplasmic reticulum membrane / glutamatergic synapse / extracellular space / plasma membrane / cytoplasm Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.15 Å MOLECULAR REPLACEMENT / Resolution: 2.15 Å | |||||||||
 Authors Authors | Geng, Y. / Bush, M. / Mosyak, L. / Wang, F. / Fan, Q.R. | |||||||||
 Citation Citation |  Journal: Nature / Year: 2013 Journal: Nature / Year: 2013Title: Structural mechanism of ligand activation in human GABA(B) receptor. Authors: Geng, Y. / Bush, M. / Mosyak, L. / Wang, F. / Fan, Q.R. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  4mr8.cif.gz 4mr8.cif.gz | 193.5 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb4mr8.ent.gz pdb4mr8.ent.gz | 148.7 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  4mr8.json.gz 4mr8.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  4mr8_validation.pdf.gz 4mr8_validation.pdf.gz | 1.1 MB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  4mr8_full_validation.pdf.gz 4mr8_full_validation.pdf.gz | 1.1 MB | Display | |
| Data in XML |  4mr8_validation.xml.gz 4mr8_validation.xml.gz | 36.3 KB | Display | |
| Data in CIF |  4mr8_validation.cif.gz 4mr8_validation.cif.gz | 52.9 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/mr/4mr8 https://data.pdbj.org/pub/pdb/validation_reports/mr/4mr8 ftp://data.pdbj.org/pub/pdb/validation_reports/mr/4mr8 ftp://data.pdbj.org/pub/pdb/validation_reports/mr/4mr8 | HTTPS FTP |
-Related structure data
| Related structure data |  4mqeSC  4mqfC  4mr7C  4mr9C  4mrmC  4ms1C  4ms3C  4ms4C S: Starting model for refinement C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
|
- Components
Components
-Gamma-aminobutyric acid type B receptor subunit ... , 2 types, 2 molecules AB
| #1: Protein | Mass: 47645.648 Da / Num. of mol.: 1 / Fragment: extracellular domain (SEE REMARK 999) Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: GABBR1, GPRC3A / Plasmid: pFBDM / Production host: Homo sapiens (human) / Gene: GABBR1, GPRC3A / Plasmid: pFBDM / Production host:  |
|---|---|
| #2: Protein | Mass: 49127.492 Da / Num. of mol.: 1 / Fragment: extracellular domain (UNP residues 42-466) Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: GABBR2, GPR51, GPRC3B / Plasmid: pFBDM / Production host: Homo sapiens (human) / Gene: GABBR2, GPR51, GPRC3B / Plasmid: pFBDM / Production host:  |
-Sugars , 3 types, 3 molecules 
| #3: Polysaccharide | beta-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4) ...beta-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose Source method: isolated from a genetically manipulated source |
|---|---|
| #4: Polysaccharide | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta- ...2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose Source method: isolated from a genetically manipulated source |
| #5: Sugar | ChemComp-NAG / |
-Non-polymers , 2 types, 475 molecules 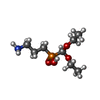


| #6: Chemical | ChemComp-2BW / ( |
|---|---|
| #7: Water | ChemComp-HOH / |
-Details
| Has protein modification | Y |
|---|---|
| Sequence details | SUBUNIT 1 IS RESIDUES 48-459 OF ISOFORM 1B (UNP Q9UBS5-2). |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 3.01 Å3/Da / Density % sol: 59.12 % |
|---|---|
| Crystal grow | Temperature: 277 K / Method: vapor diffusion, hanging drop / pH: 7 Details: 10% PEG3350, 20% glycerol, 0.12 M sodium acetate, pH 7.0, VAPOR DIFFUSION, HANGING DROP, temperature 277K |
-Data collection
| Diffraction | Mean temperature: 100 K | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  APS APS  / Beamline: 24-ID-C / Wavelength: 0.97949 Å / Beamline: 24-ID-C / Wavelength: 0.97949 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Detector | Type: ADSC QUANTUM 315 / Detector: CCD / Date: Aug 16, 2011 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Radiation | Monochromator: cryo-cooled double crystal Si(111) / Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Radiation wavelength | Wavelength: 0.97949 Å / Relative weight: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reflection | Resolution: 2.15→112.749 Å / Num. all: 61684 / Num. obs: 61684 / % possible obs: 99.2 % / Observed criterion σ(F): 0 / Observed criterion σ(I): 0 / Redundancy: 3.7 % / Biso Wilson estimate: 48.23 Å2 / Rmerge(I) obs: 0.037 / Rsym value: 0.037 / Net I/σ(I): 18.6 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reflection shell | Diffraction-ID: 1
|
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: PDB ENTRY 4MQE Resolution: 2.15→59.67 Å / Cor.coef. Fo:Fc: 0.9327 / Cor.coef. Fo:Fc free: 0.9316 / Occupancy max: 1 / Occupancy min: 1 / SU R Cruickshank DPI: 0.208 / Cross valid method: THROUGHOUT / σ(F): 0 / σ(I): 0 / SU R Blow DPI: 0.208 / SU Rfree Blow DPI: 0.161 / SU Rfree Cruickshank DPI: 0.163 / Stereochemistry target values: Engh & Huber
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso max: 143.33 Å2 / Biso mean: 53.2603 Å2 / Biso min: 14.76 Å2
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine analyze | Luzzati coordinate error obs: 0.303 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.15→59.67 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 2.15→2.21 Å / Total num. of bins used: 20
|
 Movie
Movie Controller
Controller












 PDBj
PDBj





