[English] 日本語
 Yorodumi
Yorodumi- ChemComp-4CT: (3R,4S)-1-[(4-amino-5H-pyrrolo[3,2-d]pyrimidin-7-yl)methyl]-4-{[(... -
+ Open data
Open data
- Basic information
Basic information
| Entry | 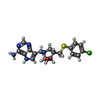 Database: PDB chemical components / ID: 4CT Database: PDB chemical components / ID: 4CT |
|---|---|
| Name | Name: ( |
-Chemical information
| Composition |  | ||||||
|---|---|---|---|---|---|---|---|
| Others | Type: NON-POLYMER / PDB classification: HETAIN / Three letter code: 4CT / Ideal coordinates details: Corina / Model coordinates PDB-ID: 3O4V | ||||||
| History |
| ||||||
 External links External links |  UniChem / UniChem /  ChemSpider / ChemSpider /  Brenda Brenda      / /  PubChem / PubChem /  PubChem_TPharma PubChem_TPharma  / /  ZINC / ZINC /  Wikipedia search / Wikipedia search /  Google search Google search |
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
-Details
-SMILES
| ACDLabs 12.01 | | CACTVS 3.370 | OpenEye OEToolkits 1.7.0 | |
|---|
-SMILES CANONICAL
| CACTVS 3.370 | | OpenEye OEToolkits 1.7.0 | |
|---|
-InChI
| InChI 1.03 |
|---|
-InChIKey
| InChI 1.03 |
|---|
-SYSTEMATIC NAME
| ACDLabs 12.01 | (| OpenEye OEToolkits 1.7.0 | ( | |
|---|
-PDB entries
Showing all 6 items

PDB-3o4v: 
Crystal structure of E. coli MTA/SAH nucleosidase in complex with (4-Chlorophenyl)thio-DADMe-ImmA

PDB-3ozc: 
Crystal Structure of human 5'-deoxy-5'-methyladenosine phosphorylase in complex with pCl-phenylthioDADMeImmA

PDB-3ozd: 
Crystal Structure of human 5'-deoxy-5'-methyladenosine phosphorylase in complex with pCl-phenylthioDADMeImmA

PDB-5k1z: 
Joint X-ray/neutron structure of MTAN complex with p-ClPh-Thio-DADMe-ImmA

PDB-5kb3: 
1.4 A resolution structure of Helicobacter Pylori MTAN in complexed with p-ClPh-DADMe-ImmA

PDB-6x6r: 
Crystal structure of C.difficile ribosyltransferase CDTa in complex with pCl-phenylthioDADMeImmA
 Movie
Movie Controller
Controller


