+ データを開く
データを開く
- 基本情報
基本情報
| 登録情報 | データベース: EMDB / ID: EMD-4933 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| タイトル | Cryo-EM structure of the microsporidian ribosome: Multibody-refined map body 3 (SSU-head) | |||||||||
 マップデータ マップデータ | Cryo-EM structure of the microsporidian ribosome: Multibody-refined map body 3 (SSU-head) | |||||||||
 試料 試料 |
| |||||||||
| 生物種 |  Vairimorpha necatrix (菌類) Vairimorpha necatrix (菌類) | |||||||||
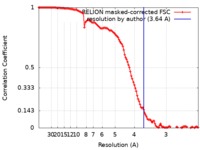
| 手法 | 単粒子再構成法 / クライオ電子顕微鏡法 / 解像度: 3.64 Å | |||||||||
 データ登録者 データ登録者 | Barandun J / Hunziker M / Vossbrinck CR / Klinge S | |||||||||
| 資金援助 |  米国, 米国,  スイス, 2件 スイス, 2件
| |||||||||
 引用 引用 |  ジャーナル: Nat Microbiol / 年: 2019 ジャーナル: Nat Microbiol / 年: 2019タイトル: Evolutionary compaction and adaptation visualized by the structure of the dormant microsporidian ribosome. 著者: Jonas Barandun / Mirjam Hunziker / Charles R Vossbrinck / Sebastian Klinge /   要旨: Microsporidia are eukaryotic parasites that infect essentially all animal species, including many of agricultural importance, and are significant opportunistic parasites of humans. They are ...Microsporidia are eukaryotic parasites that infect essentially all animal species, including many of agricultural importance, and are significant opportunistic parasites of humans. They are characterized by having a specialized infection apparatus, an obligate intracellular lifestyle, rudimentary mitochondria and the smallest known eukaryotic genomes. Extreme genome compaction led to minimal gene sizes affecting even conserved ancient complexes such as the ribosome. In the present study, the cryo-electron microscopy structure of the ribosome from the microsporidium Vairimorpha necatrix is presented, which illustrates how genome compaction has resulted in the smallest known eukaryotic cytoplasmic ribosome. Selection pressure led to the loss of two ribosomal proteins and removal of essentially all eukaryote-specific ribosomal RNA (rRNA) expansion segments, reducing the rRNA to a functionally conserved core. The structure highlights how one microsporidia-specific and several repurposed existing ribosomal proteins compensate for the extensive rRNA reduction. The microsporidian ribosome is kept in an inactive state by two previously uncharacterized dormancy factors that specifically target the functionally important E-site, P-site and polypeptide exit tunnel. The present study illustrates the distinct effects of evolutionary pressure on RNA and protein-coding genes, provides a mechanism for ribosome inhibition and can serve as a structural basis for the development of inhibitors against microsporidian parasites. | |||||||||
| 履歴 |
|
- 構造の表示
構造の表示
| ムービー |
 ムービービューア ムービービューア |
|---|---|
| 構造ビューア | EMマップ:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| 添付画像 |
- ダウンロードとリンク
ダウンロードとリンク
-EMDBアーカイブ
| マップデータ |  emd_4933.map.gz emd_4933.map.gz | 3.6 MB |  EMDBマップデータ形式 EMDBマップデータ形式 | |
|---|---|---|---|---|
| ヘッダ (付随情報) |  emd-4933-v30.xml emd-4933-v30.xml emd-4933.xml emd-4933.xml | 22.2 KB 22.2 KB | 表示 表示 |  EMDBヘッダ EMDBヘッダ |
| FSC (解像度算出) |  emd_4933_fsc.xml emd_4933_fsc.xml | 12.1 KB | 表示 |  FSCデータファイル FSCデータファイル |
| 画像 |  emd_4933.png emd_4933.png | 210.4 KB | ||
| アーカイブディレクトリ |  http://ftp.pdbj.org/pub/emdb/structures/EMD-4933 http://ftp.pdbj.org/pub/emdb/structures/EMD-4933 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4933 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4933 | HTTPS FTP |
-検証レポート
| 文書・要旨 |  emd_4933_validation.pdf.gz emd_4933_validation.pdf.gz | 237.2 KB | 表示 |  EMDB検証レポート EMDB検証レポート |
|---|---|---|---|---|
| 文書・詳細版 |  emd_4933_full_validation.pdf.gz emd_4933_full_validation.pdf.gz | 236.3 KB | 表示 | |
| XML形式データ |  emd_4933_validation.xml.gz emd_4933_validation.xml.gz | 12.6 KB | 表示 | |
| アーカイブディレクトリ |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-4933 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-4933 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-4933 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-4933 | HTTPS FTP |
-関連構造データ
| 関連構造データ |  4931C  4932C  4934C  4935C  6rm3C C: 同じ文献を引用 ( |
|---|---|
| 類似構造データ | |
| 電子顕微鏡画像生データ |  EMPIAR-11075 (タイトル: Single-particle cryo-EM dataset of the Vairimorpha necatrix ribosome EMPIAR-11075 (タイトル: Single-particle cryo-EM dataset of the Vairimorpha necatrix ribosomeData size: 1.0 TB Data #1: Unaligned multi-frame micrographs of the Vairimorpha necatrix ribosome [micrographs - multiframe]) |
- リンク
リンク
| EMDBのページ |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| 「今月の分子」の関連する項目 |
- マップ
マップ
| ファイル |  ダウンロード / ファイル: emd_4933.map.gz / 形式: CCP4 / 大きさ: 149.9 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) ダウンロード / ファイル: emd_4933.map.gz / 形式: CCP4 / 大きさ: 149.9 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 注釈 | Cryo-EM structure of the microsporidian ribosome: Multibody-refined map body 3 (SSU-head) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 投影像・断面図 | 画像のコントロール
画像は Spider により作成 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ボクセルのサイズ | X=Y=Z: 1.2 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
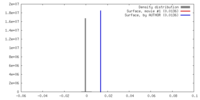
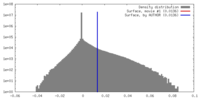
| 密度 |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 対称性 | 空間群: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 詳細 | EMDB XML:
CCP4マップ ヘッダ情報:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-添付データ
- 試料の構成要素
試料の構成要素
-全体 : Microsporidian Ribosome
| 全体 | 名称: Microsporidian Ribosome |
|---|---|
| 要素 |
|
-超分子 #1: Microsporidian Ribosome
| 超分子 | 名称: Microsporidian Ribosome / タイプ: complex / ID: 1 / 親要素: 0 / 含まれる分子: #1-#78 |
|---|---|
| 由来(天然) | 生物種:  Vairimorpha necatrix (菌類) Vairimorpha necatrix (菌類) |
-実験情報
-構造解析
| 手法 | クライオ電子顕微鏡法 |
|---|---|
 解析 解析 | 単粒子再構成法 |
| 試料の集合状態 | particle |
- 試料調製
試料調製
| 濃度 | 11 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 緩衝液 | pH: 7.5 構成要素:
| ||||||||||||
| 凍結 | 凍結剤: ETHANE / チャンバー内湿度: 100 % / チャンバー内温度: 295 K / 装置: FEI VITROBOT MARK IV |
- 電子顕微鏡法
電子顕微鏡法
| 顕微鏡 | FEI TALOS ARCTICA |
|---|---|
| 撮影 | フィルム・検出器のモデル: GATAN K2 SUMMIT (4k x 4k) 検出モード: SUPER-RESOLUTION / 撮影したグリッド数: 2 / 実像数: 3284 / 平均露光時間: 0.25 sec. / 平均電子線量: 5.55 e/Å2 |
| 電子線 | 加速電圧: 200 kV / 電子線源:  FIELD EMISSION GUN FIELD EMISSION GUN |
| 電子光学系 | 最小 デフォーカス(補正後): 4.0 µm / 照射モード: FLOOD BEAM / 撮影モード: BRIGHT FIELD / 最小 デフォーカス(公称値): 0.5 µm |
| 試料ステージ | 試料ホルダーモデル: FEI TITAN KRIOS AUTOGRID HOLDER ホルダー冷却材: NITROGEN |
| 実験機器 |  モデル: Talos Arctica / 画像提供: FEI Company |
+ 画像解析
画像解析
-原子モデル構築 1
| 初期モデル | PDB ID: |
|---|---|
| 精密化 | 空間: REAL / プロトコル: OTHER |
 ムービー
ムービー コントローラー
コントローラー













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)