[English] 日本語
 Yorodumi
Yorodumi- EMDB-4889: Structure of Vaccinia Virus DNA-dependent RNA polymerase elongati... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-4889 | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of Vaccinia Virus DNA-dependent RNA polymerase elongation complex | ||||||||||||||||||||||||
 Map data Map data | |||||||||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||||||||
 Keywords Keywords | Vaccinia / RNA polymerase / Transcription / Gene expression / VIRAL PROTEIN | ||||||||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationvirion component => GO:0044423 / viral transcription / ribonucleoside binding / DNA-directed 5'-3' RNA polymerase activity / DNA-directed RNA polymerase / DNA-templated transcription / DNA binding / zinc ion binding Similarity search - Function | ||||||||||||||||||||||||
| Biological species |  Vaccinia virus GLV-1h68 / Vaccinia virus GLV-1h68 /  | ||||||||||||||||||||||||
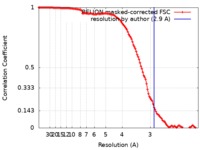
| Method | single particle reconstruction / cryo EM / Resolution: 2.9 Å | ||||||||||||||||||||||||
 Authors Authors | Hillen HS / Bartuli J | ||||||||||||||||||||||||
| Funding support |  Germany, 7 items Germany, 7 items
| ||||||||||||||||||||||||
 Citation Citation |  Journal: Cell / Year: 2019 Journal: Cell / Year: 2019Title: Structural Basis of Poxvirus Transcription: Transcribing and Capping Vaccinia Complexes. Authors: Hauke S Hillen / Julia Bartuli / Clemens Grimm / Christian Dienemann / Kristina Bedenk / Aladar A Szalay / Utz Fischer / Patrick Cramer /   Abstract: Poxviruses use virus-encoded multisubunit RNA polymerases (vRNAPs) and RNA-processing factors to generate mG-capped mRNAs in the host cytoplasm. In the accompanying paper, we report structures of ...Poxviruses use virus-encoded multisubunit RNA polymerases (vRNAPs) and RNA-processing factors to generate mG-capped mRNAs in the host cytoplasm. In the accompanying paper, we report structures of core and complete vRNAP complexes of the prototypic Vaccinia poxvirus (Grimm et al., 2019; in this issue of Cell). Here, we present the cryo-electron microscopy (cryo-EM) structures of Vaccinia vRNAP in the form of a transcribing elongation complex and in the form of a co-transcriptional capping complex that contains the viral capping enzyme (CE). The trifunctional CE forms two mobile modules that bind the polymerase surface around the RNA exit tunnel. RNA extends from the vRNAP active site through this tunnel and into the active site of the CE triphosphatase. Structural comparisons suggest that growing RNA triggers large-scale rearrangements on the surface of the transcription machinery during the transition from transcription initiation to RNA capping and elongation. Our structures unravel the basis for synthesis and co-transcriptional modification of poxvirus RNA. | ||||||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
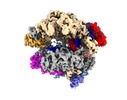
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_4889.map.gz emd_4889.map.gz | 10.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-4889-v30.xml emd-4889-v30.xml emd-4889.xml emd-4889.xml | 36.7 KB 36.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_4889_fsc.xml emd_4889_fsc.xml | 12 KB | Display |  FSC data file FSC data file |
| Images |  emd_4889.png emd_4889.png | 140.7 KB | ||
| Masks |  emd_4889_msk_1.map emd_4889_msk_1.map | 149.9 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-4889.cif.gz emd-4889.cif.gz | 9.4 KB | ||
| Others |  emd_4889_additional.map.gz emd_4889_additional.map.gz emd_4889_half_map_1.map.gz emd_4889_half_map_1.map.gz emd_4889_half_map_2.map.gz emd_4889_half_map_2.map.gz | 118.6 MB 118.5 MB 118.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-4889 http://ftp.pdbj.org/pub/emdb/structures/EMD-4889 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4889 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4889 | HTTPS FTP |
-Validation report
| Summary document |  emd_4889_validation.pdf.gz emd_4889_validation.pdf.gz | 849.6 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_4889_full_validation.pdf.gz emd_4889_full_validation.pdf.gz | 849.2 KB | Display | |
| Data in XML |  emd_4889_validation.xml.gz emd_4889_validation.xml.gz | 19.3 KB | Display | |
| Data in CIF |  emd_4889_validation.cif.gz emd_4889_validation.cif.gz | 25.4 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-4889 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-4889 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-4889 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-4889 | HTTPS FTP |
-Related structure data
| Related structure data |  6ridMC  4888C  4890C  4891C  6ricC  6rieC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_4889.map.gz / Format: CCP4 / Size: 149.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_4889.map.gz / Format: CCP4 / Size: 149.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
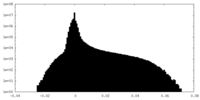
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.05 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
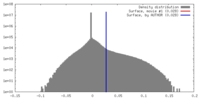
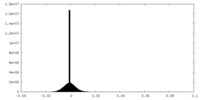
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
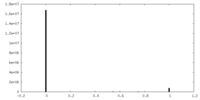
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_4889_msk_1.map emd_4889_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: #1
| File | emd_4889_additional.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_4889_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_4889_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
+Entire : Vaccinia Virus DNA-dependent RNA polymerase elongation complex
+Supramolecule #1: Vaccinia Virus DNA-dependent RNA polymerase elongation complex
+Supramolecule #2: DNA-directed RNA polymerase subunits
+Supramolecule #3: DNA-dependent RNA polymerase subunit rpo132
+Supramolecule #4: Nucleic acids
+Macromolecule #1: DNA-dependent RNA polymerase subunit rpo147
+Macromolecule #2: DNA-dependent RNA polymerase subunit rpo132
+Macromolecule #3: DNA-directed RNA polymerase 35 kDa subunit
+Macromolecule #4: DNA-directed RNA polymerase subunit
+Macromolecule #5: DNA-directed RNA polymerase 19 kDa subunit
+Macromolecule #6: DNA-dependent RNA polymerase subunit rpo18
+Macromolecule #7: DNA-dependent RNA polymerase subunit rpo7
+Macromolecule #8: DNA-directed RNA polymerase 30 kDa polypeptide
+Macromolecule #9: Non-template strand DNA
+Macromolecule #10: Template strand DNA
+Macromolecule #11: RNA
+Macromolecule #12: MAGNESIUM ION
+Macromolecule #13: ZINC ION
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK II |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 QUANTUM (4k x 4k) / Detector mode: COUNTING / Number grids imaged: 1 / Number real images: 4958 / Average electron dose: 1.02 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal magnification: 130000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: AB INITIO MODEL |
|---|---|
| Output model |  PDB-6rid: |
 Movie
Movie Controller
Controller















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)