[English] 日本語
 Yorodumi
Yorodumi- EMDB-4747: CryoEM structure of calcium-bound human TMEM16K / Anoctamin 10 in... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-4747 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | CryoEM structure of calcium-bound human TMEM16K / Anoctamin 10 in detergent (low Ca2+, closed form) | |||||||||
 Map data Map data | RELION3 postprocessed map sharpened with a bfactor of -176A**2 | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | MEMBRANE PROTEIN / CALCIUM ACTIVATION / TRANSPORT PROTEIN / LIPID TRANSPORT / Structural Genomics / Structural Genomics Consortium / SGC | |||||||||
| Function / homology |  Function and homology information Function and homology informationintracellularly calcium-gated chloride channel activity / calcium-activated cation channel activity / chloride channel activity / chloride transmembrane transport / Stimuli-sensing channels / monoatomic ion transmembrane transport / Induction of Cell-Cell Fusion / cilium / intracellular membrane-bounded organelle / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
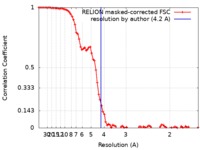
| Method | single particle reconstruction / cryo EM / Resolution: 4.2 Å | |||||||||
 Authors Authors | Pike ACW / Bushell SR | |||||||||
| Funding support |  United Kingdom, 2 items United Kingdom, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2019 Journal: Nat Commun / Year: 2019Title: The structural basis of lipid scrambling and inactivation in the endoplasmic reticulum scramblase TMEM16K. Authors: Simon R Bushell / Ashley C W Pike / Maria E Falzone / Nils J G Rorsman / Chau M Ta / Robin A Corey / Thomas D Newport / John C Christianson / Lara F Scofano / Chitra A Shintre / Annamaria ...Authors: Simon R Bushell / Ashley C W Pike / Maria E Falzone / Nils J G Rorsman / Chau M Ta / Robin A Corey / Thomas D Newport / John C Christianson / Lara F Scofano / Chitra A Shintre / Annamaria Tessitore / Amy Chu / Qinrui Wang / Leela Shrestha / Shubhashish M M Mukhopadhyay / James D Love / Nicola A Burgess-Brown / Rebecca Sitsapesan / Phillip J Stansfeld / Juha T Huiskonen / Paolo Tammaro / Alessio Accardi / Elisabeth P Carpenter /    Abstract: Membranes in cells have defined distributions of lipids in each leaflet, controlled by lipid scramblases and flip/floppases. However, for some intracellular membranes such as the endoplasmic ...Membranes in cells have defined distributions of lipids in each leaflet, controlled by lipid scramblases and flip/floppases. However, for some intracellular membranes such as the endoplasmic reticulum (ER) the scramblases have not been identified. Members of the TMEM16 family have either lipid scramblase or chloride channel activity. Although TMEM16K is widely distributed and associated with the neurological disorder autosomal recessive spinocerebellar ataxia type 10 (SCAR10), its location in cells, function and structure are largely uncharacterised. Here we show that TMEM16K is an ER-resident lipid scramblase with a requirement for short chain lipids and calcium for robust activity. Crystal structures of TMEM16K show a scramblase fold, with an open lipid transporting groove. Additional cryo-EM structures reveal extensive conformational changes from the cytoplasmic to the ER side of the membrane, giving a state with a closed lipid permeation pathway. Molecular dynamics simulations showed that the open-groove conformation is necessary for scramblase activity. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_4747.map.gz emd_4747.map.gz | 41.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-4747-v30.xml emd-4747-v30.xml emd-4747.xml emd-4747.xml | 21.4 KB 21.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_4747_fsc.xml emd_4747_fsc.xml | 8.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_4747.png emd_4747.png | 178.8 KB | ||
| Masks |  emd_4747_msk_1.map emd_4747_msk_1.map | 45.2 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-4747.cif.gz emd-4747.cif.gz | 7.1 KB | ||
| Others |  emd_4747_half_map_1.map.gz emd_4747_half_map_1.map.gz emd_4747_half_map_2.map.gz emd_4747_half_map_2.map.gz | 33.7 MB 33.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-4747 http://ftp.pdbj.org/pub/emdb/structures/EMD-4747 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4747 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4747 | HTTPS FTP |
-Related structure data
| Related structure data |  6r7yMC  4746C  4748C  5oc9C  6r65C  6r7xC  6r7zC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_4747.map.gz / Format: CCP4 / Size: 45.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_4747.map.gz / Format: CCP4 / Size: 45.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | RELION3 postprocessed map sharpened with a bfactor of -176A**2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.822 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_4747_msk_1.map emd_4747_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: RELION3 halfmap1
| File | emd_4747_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | RELION3 halfmap1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: RELION3 halfmap2
| File | emd_4747_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | RELION3 halfmap2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Anoctamin 10
| Entire | Name: Anoctamin 10 |
|---|---|
| Components |
|
-Supramolecule #1: Anoctamin 10
| Supramolecule | Name: Anoctamin 10 / type: organelle_or_cellular_component / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 153 KDa |
-Macromolecule #1: Anoctamin-10
| Macromolecule | Name: Anoctamin-10 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 77.276016 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MKVTLSALDT SESSFTPLVV IELAQDVKEE TKEWLKNRII AKKKDGGAQL LFRPLLNKYE QETLENQNLY LVGASKIRML LGAEAVGLV KECNDNTMRA FTYRTRQNFK GFDDNNDDFL TMAECQFIIK HELENLRAKD EKMIPGYPQA KLYPGKSLLR R LLTSGIVI ...String: MKVTLSALDT SESSFTPLVV IELAQDVKEE TKEWLKNRII AKKKDGGAQL LFRPLLNKYE QETLENQNLY LVGASKIRML LGAEAVGLV KECNDNTMRA FTYRTRQNFK GFDDNNDDFL TMAECQFIIK HELENLRAKD EKMIPGYPQA KLYPGKSLLR R LLTSGIVI QVFPLHDSEA LKKLEDTWYT RFALKYQPID SIRGYFGETI ALYFGFLEYF TFALIPMAVI GLPYYLFVWE DY DKYVIFA SFNLIWSTVI LELWKRGCAN MTYRWGTLLM KRKFEEPRPG FHGVLGINSI TGKEEPLYPS YKRQLRIYLV SLP FVCLCL YFSLYVMMIY FDMEVWALGL HENSGSEWTS VLLYVPSIIY AIVIEIMNRL YRYAAEFLTS WENHRLESAY QNHL ILKVL VFNFLNCFAS LFYIAFVLKD MKLLRQSLAT LLITSQILNQ IMESFLPYWL QRKHGVRVKR KVQALKADID ATLYE QVIL EKEMGTYLGT FDDYLELFLQ FGYVSLFSCV YPLAAAFAVL NNFTEVNSDA LKMCRVFKRP FSEPSANIGV WQLAFE TMS VISVVTNCAL IGMSPQVNAV FPESKADLIL IVVAVEHALL ALKFILAFAI PDKPRHIQMK LARLEFESLE ALKQQQM KL VTENLKEEPM ESGKEKATAE NLYFQ UniProtKB: Anoctamin-10 |
-Macromolecule #2: CALCIUM ION
| Macromolecule | Name: CALCIUM ION / type: ligand / ID: 2 / Number of copies: 6 / Formula: CA |
|---|---|
| Molecular weight | Theoretical: 40.078 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 3 mg/mL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
Details: No additional calcium added. Background levels of calcium estimated to be ca. 500nM | |||||||||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. / Pretreatment - Atmosphere: AIR | |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 278 K / Instrument: FEI VITROBOT MARK IV / Details: blot for 4.5sec prior to plunging. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Frames/image: 1-20 / Number grids imaged: 1 / Number real images: 1975 / Average exposure time: 8.0 sec. / Average electron dose: 56.5 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.75 µm / Nominal defocus min: 1.0 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Details | Refined using phenix.real_space_refine with NCS constraints and model reference restraints used (PDB id: TBC) against RELION3 post-processed map. |
|---|---|
| Refinement | Space: REAL / Protocol: AB INITIO MODEL / Overall B value: 176 / Target criteria: Correlation coef |
| Output model |  PDB-6r7y: |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)