+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of HMPV (MPV-2cREKR) | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Prefusion / HMPV / Cryo-EM / VIRUS / VIRAL PROTEIN | |||||||||
| Biological species |  Human metapneumovirus Human metapneumovirus | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.14 Å | |||||||||
 Authors Authors | Yu X / Langedijk JPM | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: Efficacious human metapneumovirus vaccine based on AI-guided engineering of a closed prefusion trimer Authors: Bakkers MJG / Ritschel T / Tiemessen M / Dijkman J / Zuffiano AA / Yu X / Overveld D / Le L / Voorzaat R / Haaren M / Man M / Sharma S / Tamara S / Fits L / Zahn R / Juraszek J / Langedijk JPM | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_43516.map.gz emd_43516.map.gz | 59.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-43516-v30.xml emd-43516-v30.xml emd-43516.xml emd-43516.xml | 13.5 KB 13.5 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_43516.png emd_43516.png | 53.5 KB | ||
| Filedesc metadata |  emd-43516.cif.gz emd-43516.cif.gz | 5.5 KB | ||
| Others |  emd_43516_half_map_1.map.gz emd_43516_half_map_1.map.gz emd_43516_half_map_2.map.gz emd_43516_half_map_2.map.gz | 59.4 MB 59.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-43516 http://ftp.pdbj.org/pub/emdb/structures/EMD-43516 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-43516 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-43516 | HTTPS FTP |
-Validation report
| Summary document |  emd_43516_validation.pdf.gz emd_43516_validation.pdf.gz | 1.1 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_43516_full_validation.pdf.gz emd_43516_full_validation.pdf.gz | 1.1 MB | Display | |
| Data in XML |  emd_43516_validation.xml.gz emd_43516_validation.xml.gz | 12.3 KB | Display | |
| Data in CIF |  emd_43516_validation.cif.gz emd_43516_validation.cif.gz | 14.4 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-43516 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-43516 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-43516 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-43516 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_43516.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_43516.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.948 Å | ||||||||||||||||||||||||||||||||||||
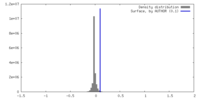
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_43516_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
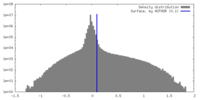
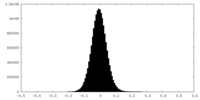
| Density Histograms |
-Half map: #2
| File | emd_43516_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
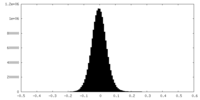
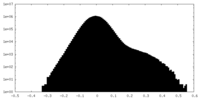
| Density Histograms |
- Sample components
Sample components
-Entire : Prefusion of HMPV (MPV-2cREKR)
| Entire | Name: Prefusion of HMPV (MPV-2cREKR) |
|---|---|
| Components |
|
-Supramolecule #1: Prefusion of HMPV (MPV-2cREKR)
| Supramolecule | Name: Prefusion of HMPV (MPV-2cREKR) / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Human metapneumovirus Human metapneumovirus |
-Macromolecule #1: Prefusion of HMPV (MPV-2c)
| Macromolecule | Name: Prefusion of HMPV (MPV-2c) / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Human metapneumovirus Human metapneumovirus |
| Molecular weight | Theoretical: 56.553586 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MSWKVVIIFS LLITPQHGLK ESYLEESCST ITEGYLSVLR TGWYTNVFTL EVGDVENLTC SDGPSLIKTE LDLTKSALRE LKTVSADQL RRRRELPRFM NYTLNNAKKT NVTLSKKRKR RFVLGAIALG RATAAAVTAG VAIAKTIRLE SEVTAIKNAL K TTNEAVST ...String: MSWKVVIIFS LLITPQHGLK ESYLEESCST ITEGYLSVLR TGWYTNVFTL EVGDVENLTC SDGPSLIKTE LDLTKSALRE LKTVSADQL RRRRELPRFM NYTLNNAKKT NVTLSKKRKR RFVLGAIALG RATAAAVTAG VAIAKTIRLE SEVTAIKNAL K TTNEAVST LGNGVRVLAT AVRELKDFVS KNLTRAINKN KCDIDDLKMA VSFSQFNRRF LNVVRQFSEN AGITPAISLD LM TDAELAR AISNMPTSAG QIKLMLENRA MVRRKGFGIL IGVYGSSVIY MVQLPIFGVI DTPCWIVKAA PSCSEKKGNY ACL LREDQG WYCQNAGSTV YYPNEKDCET RGDHVFCDTA AGINVAEQSK ECNINISTTN YPCKVSTGRN PISMVALSPL GALV ACYKG VSCSIGSNRV GIIKQLNKGC SYITNQDADT VTIDNTVYQL SKVEGEQHVI KGRPVSSSFD PIKFPPDQFN VALDQ VFEN IENSQAWVRK FDEILSSIEK GNTGGSEPEA |
-Macromolecule #4: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 4 / Number of copies: 3 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.4 µm / Nominal defocus min: 1.2 µm |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: INSILICO MODEL |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.14 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 97386 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)