+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | ssRNA phage PhiCb5 virion | |||||||||
 Map data Map data | map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | ssRNA phage / VIRUS | |||||||||
| Function / homology | Phage phiCb5, coat protein / Assembly protein / Phage maturation protein / virion attachment to host cell pilus / virion component / viral capsid / Maturation protein / Coat protein Function and homology information Function and homology information | |||||||||
| Biological species |  Caulobacter vibrioides (bacteria) / Caulobacter vibrioides (bacteria) /  Caulobacter phage phiCb5 (virus) Caulobacter phage phiCb5 (virus) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.7 Å | |||||||||
 Authors Authors | Wang Y / Zhang J | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Sci Adv / Year: 2024 Journal: Sci Adv / Year: 2024Title: Structural mechanisms of Tad pilus assembly and its interaction with an RNA virus. Authors: Yuhang Wang / Matthew Theodore / Zhongliang Xing / Utkarsh Narsaria / Zihao Yu / Lanying Zeng / Junjie Zhang /  Abstract: Tad (tight adherence) pili, part of the type IV pili family, are crucial for mechanosensing, surface adherence, bacteriophage (phage) adsorption, and cell-cycle regulation. Unlike other type IV ... Tad (tight adherence) pili, part of the type IV pili family, are crucial for mechanosensing, surface adherence, bacteriophage (phage) adsorption, and cell-cycle regulation. Unlike other type IV pilins, Tad pilins lack the typical globular β sheet domain responsible for pilus assembly and phage binding. The mechanisms of Tad pilus assembly and its interaction with phage ΦCb5 have been elusive. Using cryo-electron microscopy, we unveiled the Tad pilus assembly mechanism, featuring a unique network of hydrogen bonds at its core. We then identified the Tad pilus binding to the ΦCb5 maturation protein (Mat) through its β region. Notably, the amino terminus of ΦCb5 Mat is exposed outside the capsid and phage/pilus interface, enabling the attachment of fluorescent and affinity tags. These engineered ΦCb5 virions can be efficiently assembled and purified in , maintaining infectivity against , which presents promising applications, including RNA delivery and phage display. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_42163.map.gz emd_42163.map.gz | 456.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-42163-v30.xml emd-42163-v30.xml emd-42163.xml emd-42163.xml | 15.9 KB 15.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_42163_fsc.xml emd_42163_fsc.xml | 16.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_42163.png emd_42163.png | 68.4 KB | ||
| Filedesc metadata |  emd-42163.cif.gz emd-42163.cif.gz | 5.5 KB | ||
| Others |  emd_42163_half_map_1.map.gz emd_42163_half_map_1.map.gz emd_42163_half_map_2.map.gz emd_42163_half_map_2.map.gz | 475.3 MB 475.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-42163 http://ftp.pdbj.org/pub/emdb/structures/EMD-42163 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-42163 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-42163 | HTTPS FTP |
-Validation report
| Summary document |  emd_42163_validation.pdf.gz emd_42163_validation.pdf.gz | 1 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_42163_full_validation.pdf.gz emd_42163_full_validation.pdf.gz | 1 MB | Display | |
| Data in XML |  emd_42163_validation.xml.gz emd_42163_validation.xml.gz | 26.4 KB | Display | |
| Data in CIF |  emd_42163_validation.cif.gz emd_42163_validation.cif.gz | 34.8 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-42163 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-42163 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-42163 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-42163 | HTTPS FTP |
-Related structure data
| Related structure data |  8uejMC  8u2bC  8ucrC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_42163.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_42163.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.86 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: half
| File | emd_42163_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half | ||||||||||||
| Projections & Slices |
| ||||||||||||
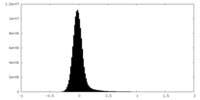
| Density Histograms |
-Half map: half
| File | emd_42163_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half | ||||||||||||
| Projections & Slices |
| ||||||||||||
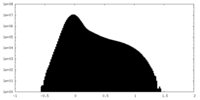
| Density Histograms |
- Sample components
Sample components
-Entire : A complex of phiCb5 maturation protein and phiCb5 shell
| Entire | Name: A complex of phiCb5 maturation protein and phiCb5 shell |
|---|---|
| Components |
|
-Supramolecule #1: A complex of phiCb5 maturation protein and phiCb5 shell
| Supramolecule | Name: A complex of phiCb5 maturation protein and phiCb5 shell type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 Details: A complex of phiCb5 maturation protein and phiCb5 shell |
|---|---|
| Source (natural) | Organism:  Caulobacter vibrioides (bacteria) Caulobacter vibrioides (bacteria) |
| Molecular weight | Theoretical: 2.1 MDa |
-Macromolecule #1: Coat protein
| Macromolecule | Name: Coat protein / type: protein_or_peptide / ID: 1 / Number of copies: 178 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Caulobacter phage phiCb5 (virus) Caulobacter phage phiCb5 (virus) |
| Molecular weight | Theoretical: 13.498981 KDa |
| Sequence | String: ALGDTLTITL GGSGGTAKVL RKINQDGYTS EYYLPETSSS FRAKVRHTKE SVKPNQVQYE RHNVEFTETV YASGSTPEFV RQAYVVIRH KVGDVSATVS DLGEALSFYL NEALYGKLIG WES UniProtKB: Coat protein |
-Macromolecule #2: Maturation protein
| Macromolecule | Name: Maturation protein / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Caulobacter phage phiCb5 (virus) Caulobacter phage phiCb5 (virus) |
| Molecular weight | Theoretical: 40.725336 KDa |
| Sequence | String: MARIRNRSSI ASSGMSTFYL FGTPIVNEEI IVRNTEWCSD VIGNPGDNPL DIHKQEWTIK PLSGQIIFGS GTYRSLQCPP EYCRGASLS HLSLPSQSGL GTTALARTNP SRPAFNLPAF IGELRDLPRM FKIAGDTMLR KGANAFLSYQ FGWKPLISDI S KALDFSAT ...String: MARIRNRSSI ASSGMSTFYL FGTPIVNEEI IVRNTEWCSD VIGNPGDNPL DIHKQEWTIK PLSGQIIFGS GTYRSLQCPP EYCRGASLS HLSLPSQSGL GTTALARTNP SRPAFNLPAF IGELRDLPRM FKIAGDTMLR KGANAFLSYQ FGWKPLISDI S KALDFSAT VRTRSDEWHR LYSNGGLKRR INLGVDIEQK KENDVVLHSS NGFVVASHTV ITVRKTWATV RWRPDAGSLP PI TKSSSEK HARALLGLGV GGLIEGAWQL MPWSWMVDWF GNVGTFLQAS NNTIGASPGL VNIMTTTTTN HQFSVKRDLS DGW IKGGDC SATVTSKARS QSSGPTITAS IPNLSGRQLS ILGALGIQRV PRHLLR UniProtKB: Maturation protein |
-Macromolecule #3: CALCIUM ION
| Macromolecule | Name: CALCIUM ION / type: ligand / ID: 3 / Number of copies: 236 / Formula: CA |
|---|---|
| Molecular weight | Theoretical: 40.078 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 Component:
Details: 20mM tris, 2mM MgCl2, 3mM CaCl2 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Grid | Model: C-flat-2/1 | ||||||||||||
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 25.0 µm / Nominal defocus min: 5.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)