+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Ghanaian virus fusion glycoprotein (GhV F) | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | GhV F / Glycoprotein / Structural Genomics / Seattle Structural Genomics Center for Infectious Disease / SSGCID / Inhibitor / VIRAL PROTEIN-IMMUNE SYSTEM complex / VIRAL PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationlyase activity / fusion of virus membrane with host plasma membrane / viral envelope / host cell plasma membrane / virion membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Henipavirus ghanaense / Henipavirus ghanaense /   Thermotoga maritima MSB8 (bacteria) Thermotoga maritima MSB8 (bacteria) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.9 Å | |||||||||
 Authors Authors | Park YJ / Seattle Structural Genomics Center for Infectious Disease (SSGCID) / Veesler D | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2024 Journal: Proc Natl Acad Sci U S A / Year: 2024Title: Structure and design of Langya virus glycoprotein antigens. Authors: Zhaoqian Wang / Matthew McCallum / Lianying Yan / Cecily A Gibson / William Sharkey / Young-Jun Park / Ha V Dang / Moushimi Amaya / Ashley Person / Christopher C Broder / David Veesler /  Abstract: Langya virus (LayV) is a recently discovered henipavirus (HNV), isolated from febrile patients in China. HNV entry into host cells is mediated by the attachment (G) and fusion (F) glycoproteins which ...Langya virus (LayV) is a recently discovered henipavirus (HNV), isolated from febrile patients in China. HNV entry into host cells is mediated by the attachment (G) and fusion (F) glycoproteins which are the main targets of neutralizing antibodies. We show here that the LayV F and G glycoproteins promote membrane fusion with human, mouse, and hamster target cells using a different, yet unknown, receptor than Nipah virus (NiV) and Hendra virus (HeV) and that NiV- and HeV-elicited monoclonal and polyclonal antibodies do not cross-react with LayV F and G. We determined cryoelectron microscopy structures of LayV F, in the prefusion and postfusion states, and of LayV G, revealing their conformational landscape and distinct antigenicity relative to NiV and HeV. We computationally designed stabilized LayV G constructs and demonstrate the generalizability of an HNV F prefusion-stabilization strategy. Our data will support the development of vaccines and therapeutics against LayV and closely related HNVs. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_41636.map.gz emd_41636.map.gz | 63.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-41636-v30.xml emd-41636-v30.xml emd-41636.xml emd-41636.xml | 17.6 KB 17.6 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_41636.png emd_41636.png | 87.9 KB | ||
| Filedesc metadata |  emd-41636.cif.gz emd-41636.cif.gz | 6.3 KB | ||
| Others |  emd_41636_additional_1.map.gz emd_41636_additional_1.map.gz emd_41636_half_map_1.map.gz emd_41636_half_map_1.map.gz emd_41636_half_map_2.map.gz emd_41636_half_map_2.map.gz | 33.8 MB 62.2 MB 62.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-41636 http://ftp.pdbj.org/pub/emdb/structures/EMD-41636 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-41636 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-41636 | HTTPS FTP |
-Validation report
| Summary document |  emd_41636_validation.pdf.gz emd_41636_validation.pdf.gz | 907.6 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_41636_full_validation.pdf.gz emd_41636_full_validation.pdf.gz | 907.2 KB | Display | |
| Data in XML |  emd_41636_validation.xml.gz emd_41636_validation.xml.gz | 11.3 KB | Display | |
| Data in CIF |  emd_41636_validation.cif.gz emd_41636_validation.cif.gz | 13.2 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-41636 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-41636 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-41636 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-41636 | HTTPS FTP |
-Related structure data
| Related structure data |  8tvbMC  8tveC  8tvfC  8tvgC  8tvhC  8tviC  8vwpC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_41636.map.gz / Format: CCP4 / Size: 67 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_41636.map.gz / Format: CCP4 / Size: 67 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1 Å | ||||||||||||||||||||||||||||||||||||
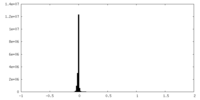
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: #1
| File | emd_41636_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
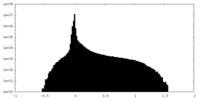
| Density Histograms |
-Half map: #2
| File | emd_41636_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
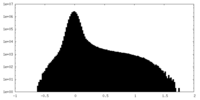
| Density Histograms |
-Half map: #1
| File | emd_41636_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
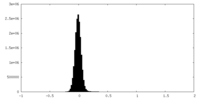
| Density Histograms |
- Sample components
Sample components
-Entire : Prefusion Ghanian henipavirus F fused to the I53-50A trimer via a...
| Entire | Name: Prefusion Ghanian henipavirus F fused to the I53-50A trimer via an intervening 16 GS linker |
|---|---|
| Components |
|
-Supramolecule #1: Prefusion Ghanian henipavirus F fused to the I53-50A trimer via a...
| Supramolecule | Name: Prefusion Ghanian henipavirus F fused to the I53-50A trimer via an intervening 16 GS linker type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Henipavirus ghanaense Henipavirus ghanaense |
-Macromolecule #1: Fusion glycoprotein F0,2-dehydro-3-deoxyphosphogluconate aldolase...
| Macromolecule | Name: Fusion glycoprotein F0,2-dehydro-3-deoxyphosphogluconate aldolase/4-hydroxy-2-oxoglutarate aldolase type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Thermotoga maritima MSB8 (bacteria) Thermotoga maritima MSB8 (bacteria)Strain: ATCC 43589 / DSM 3109 / JCM 10099 / NBRC 100826 / MSB8 |
| Molecular weight | Theoretical: 96.991531 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MKKKTDNPTI SKRGHNHSRG IKSRALLRET DNYSNGLIVE NLVRNCHHPS KNNLNYTKTQ KRDSTIPYRV EERKGHYPKI KHLIDKSYK HIKRGKRRNG HNGNIITIIL LLILILKTQM SEGAIHYETL SKIGLIKGIT REYKVKGTPS SKDIVIKLIP N VTGLNKCT ...String: MKKKTDNPTI SKRGHNHSRG IKSRALLRET DNYSNGLIVE NLVRNCHHPS KNNLNYTKTQ KRDSTIPYRV EERKGHYPKI KHLIDKSYK HIKRGKRRNG HNGNIITIIL LLILILKTQM SEGAIHYETL SKIGLIKGIT REYKVKGTPS SKDIVIKLIP N VTGLNKCT NISMENYKEQ LDKILIPINN IIELYANSTK SAPGNARFAG VIIAGVALGV AAAAQITAGI ALHEARQNAE RI NLLKDSI SATNNAVAEL QEATGGIVNV ITGMQDYINT NLVPQIDKLQ CSQIKTALDI SLSQYYSEIL TVFGPNLQNP VTT SMSIQA ISQSFGGNID LLLNLLGYTA NDLLDLLESK SITGQITYIN LEHYFMVIRV YYPIMTTISN AYVQELIKIS FNVD GSEWV SLVPSYILIR NSYLSNIDIS ECLITKNSVI CRHDFAMPMS YTLKECLTGD TEKCPREAVV TSYVPRFAIS GGVIY ANCL STTCQCYQTG KVIAQDGSQT LMMIDNQTCS IVRIEEILIS TGKYLGSQEY NTMHVSVGNP VFTDKLDITS QISNIN QSI EQSKFYLDKS KAILDKINLN LIGMKQIEDK IEEILSKIYH IENEIARIKK LIGEAPGGIE GRGGSGSGGS GGSGSEK AA KAEEAARKME ELFKKHKIVA VLRANSVEEA IEKAVAVFAG GVHLIEITFT VPDADTVIKA LSVLKEKGAI IGAGTVTS V EQARKAVESG AEFIVSPHLD EEISQFAKEK GVFYMPGVMT PTELVKAMKL GHTILKLFPG EVVGPQFVKA MKGPFPNVK FVPTGGVNLD NVAEWFKAGV LAVGVGSALV KGTPDEVREK AKAFVEKIRG ATEGGSGGSH HHHHHGSGGG SGLNDIFEAQ KIEWHE UniProtKB: Fusion glycoprotein F0, 2-dehydro-3-deoxyphosphogluconate aldolase/4-hydroxy-2-oxoglutarate aldolase |
-Macromolecule #2: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 2 / Number of copies: 12 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 63.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: OTHER / Details: ab initio |
|---|---|
| Final reconstruction | Applied symmetry - Point group: C3 (3 fold cyclic) / Algorithm: BACK PROJECTION / Resolution.type: BY AUTHOR / Resolution: 2.9 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: cryoSPARC / Number images used: 60207 |
| Initial angle assignment | Type: PROJECTION MATCHING |
| Final angle assignment | Type: PROJECTION MATCHING |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)