[English] 日本語
 Yorodumi
Yorodumi- EMDB-43593: Langya Virus attachment (G) glycoprotein with K85L/L86K mutation -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Langya Virus attachment (G) glycoprotein with K85L/L86K mutation | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | langya / attachment / glycoprotein / Structural Genomics / Seattle Structural Genomics Center for Infectious Disease / SSGCID / VIRAL PROTEIN | |||||||||
| Biological species |  Langya virus Langya virus | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.21 Å | |||||||||
 Authors Authors | Gibson CG / McCallum MM / Veesler DV | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2024 Journal: Proc Natl Acad Sci U S A / Year: 2024Title: Structure and design of Langya virus glycoprotein antigens. Authors: Zhaoqian Wang / Matthew McCallum / Lianying Yan / Cecily A Gibson / William Sharkey / Young-Jun Park / Ha V Dang / Moushimi Amaya / Ashley Person / Christopher C Broder / David Veesler /  Abstract: Langya virus (LayV) is a recently discovered henipavirus (HNV), isolated from febrile patients in China. HNV entry into host cells is mediated by the attachment (G) and fusion (F) glycoproteins which ...Langya virus (LayV) is a recently discovered henipavirus (HNV), isolated from febrile patients in China. HNV entry into host cells is mediated by the attachment (G) and fusion (F) glycoproteins which are the main targets of neutralizing antibodies. We show here that the LayV F and G glycoproteins promote membrane fusion with human, mouse, and hamster target cells using a different, yet unknown, receptor than Nipah virus (NiV) and Hendra virus (HeV) and that NiV- and HeV-elicited monoclonal and polyclonal antibodies do not cross-react with LayV F and G. We determined cryoelectron microscopy structures of LayV F, in the prefusion and postfusion states, and of LayV G, revealing their conformational landscape and distinct antigenicity relative to NiV and HeV. We computationally designed stabilized LayV G constructs and demonstrate the generalizability of an HNV F prefusion-stabilization strategy. Our data will support the development of vaccines and therapeutics against LayV and closely related HNVs. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_43593.map.gz emd_43593.map.gz | 307 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-43593-v30.xml emd-43593-v30.xml emd-43593.xml emd-43593.xml | 16.7 KB 16.7 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_43593.png emd_43593.png | 53 KB | ||
| Filedesc metadata |  emd-43593.cif.gz emd-43593.cif.gz | 5.8 KB | ||
| Others |  emd_43593_additional_1.map.gz emd_43593_additional_1.map.gz emd_43593_half_map_1.map.gz emd_43593_half_map_1.map.gz emd_43593_half_map_2.map.gz emd_43593_half_map_2.map.gz | 162 MB 301.5 MB 301.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-43593 http://ftp.pdbj.org/pub/emdb/structures/EMD-43593 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-43593 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-43593 | HTTPS FTP |
-Validation report
| Summary document |  emd_43593_validation.pdf.gz emd_43593_validation.pdf.gz | 903.4 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_43593_full_validation.pdf.gz emd_43593_full_validation.pdf.gz | 903 KB | Display | |
| Data in XML |  emd_43593_validation.xml.gz emd_43593_validation.xml.gz | 17 KB | Display | |
| Data in CIF |  emd_43593_validation.cif.gz emd_43593_validation.cif.gz | 20.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-43593 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-43593 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-43593 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-43593 | HTTPS FTP |
-Related structure data
| Related structure data |  8vwpMC  8tvbC  8tveC  8tvfC  8tvgC  8tvhC  8tviC M: atomic model generated by this map C: citing same article ( |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_43593.map.gz / Format: CCP4 / Size: 325 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_43593.map.gz / Format: CCP4 / Size: 325 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.89 Å | ||||||||||||||||||||||||||||||||||||
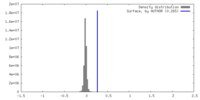
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: #1
| File | emd_43593_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
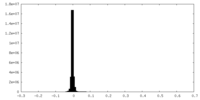
| Density Histograms |
-Half map: #1
| File | emd_43593_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
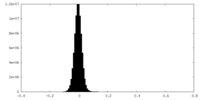
| Density Histograms |
-Half map: #2
| File | emd_43593_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
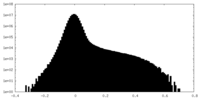
| Density Histograms |
- Sample components
Sample components
-Entire : Langya virus attachment (G) protein
| Entire | Name: Langya virus attachment (G) protein |
|---|---|
| Components |
|
-Supramolecule #1: Langya virus attachment (G) protein
| Supramolecule | Name: Langya virus attachment (G) protein / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Langya virus Langya virus |
-Macromolecule #1: Langya virus attachment (G) protein
| Macromolecule | Name: Langya virus attachment (G) protein / type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Langya virus Langya virus |
| Molecular weight | Theoretical: 59.402613 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: LKAIQAMLKI IQDEVNSLKE MLVSLDQLVK TEIKPKVSLI NTAVSVSIPA QISNLQTKVL QKLVYLEESI TKQCT(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) ...String: LKAIQAMLKI IQDEVNSLKE MLVSLDQLVK TEIKPKVSLI NTAVSVSIPA QISNLQTKVL QKLVYLEESI TKQCT(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) DTTDDDKVDT TIKPVEYYKP DGCNKTNDHF TMQPGVNFYT VPNLGPSSSS ADECYTNPS FSIGSSIYMF SQEIRKTDCT TGEILSIQIV LGRIVDKGQQ GPQASPLLVW SVPNPKIINS CAVAAGDETG W VLCSVTLT AASGEPIPHM FDGFWLYKFE PDTEVVAYRI TGFAYLLDKV YDSVFIGKGG GIQRGNDLYF QMFGLSRNRQ SI KALCEHG SCL(UNK)(UNK)(UNK)(UNK)GGY QVLCDRAVMS FGSEESLISN AYLKVNDVAS GKPTIISQTF PPSDSYK GS NGRIYTIGER YGIYLAPSSW NRYLRFGLTP DISVRSTTWL KEKDPIMKVL TTCTNTDKDM CPEICNTRGY QDIFPLSE D SSFYTYIGIT PSNEGTKSFV AVKDDAGHVA SITILPNYYS ITSATISCFM YKEEIWCIAV TEGRKQKENP QRIYAHSYR VQKMCFNI |
-Macromolecule #2: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 2 / Number of copies: 4 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.8 |
|---|---|
| Grid | Model: UltrAuFoil / Material: GOLD |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS GLACIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 47.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.5 µm |
- Image processing
Image processing
| Startup model | Type of model: OTHER / Details: CryoSPARC ab initio |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.21 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: cryoSPARC / Number images used: 79301 |
| Initial angle assignment | Type: PROJECTION MATCHING |
| Final angle assignment | Type: PROJECTION MATCHING |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)