[English] 日本語
 Yorodumi
Yorodumi- EMDB-38672: Icosahedral Reconstructed map of Car4-bound state AAV9P31 at 1.76... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
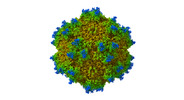
| Title | Icosahedral Reconstructed map of Car4-bound state AAV9P31 at 1.76 Angstroms | |||||||||
 Map data Map data | ||||||||||
 Sample Sample | Adeno-associated virus != Adeno-associated virus 9 Adeno-associated virus
| |||||||||
 Keywords Keywords | Adeno-associated virus 9P31 / complex / Cryo-EM / VIRUS | |||||||||
| Biological species |   Adeno-associated virus 9 Adeno-associated virus 9 | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 1.76 Å | |||||||||
 Authors Authors | Zhang R / Liu Y / Yu F / Xu G / Li L / Li B / Lou Z | |||||||||
| Funding support |  China, 2 items China, 2 items
| |||||||||
 Citation Citation |  Journal: PLoS Pathog / Year: 2024 Journal: PLoS Pathog / Year: 2024Title: Structural basis of the recognition of adeno-associated virus by the neurological system-related receptor carbonic anhydrase IV. Authors: Ran Zhang / Yixiao Liu / Fengxi Yu / Guangxue Xu / Lili Li / Baobin Li / Zhiyong Lou /   Abstract: Carbonic anhydrase IV (Car4) is a newly identified receptor that allows adeno-associated virus (AAV) 9P31 to cross the blood-brain barrier and achieve efficient infection in the central nervous ...Carbonic anhydrase IV (Car4) is a newly identified receptor that allows adeno-associated virus (AAV) 9P31 to cross the blood-brain barrier and achieve efficient infection in the central nervous system (CNS) in mouse models. However, the molecular mechanism by which engineered AAV capsids with 7-mer insertion in the variable region (VR) VIII recognize these novel cellular receptors is unknown. Here we report the cryo-EM structures of AAV9P31 and its complex with Mus musculus Car4 at atomic resolution by utilizing the block-based reconstruction (BBR) method. The structures demonstrated that Car4 binds to the protrusions at 3-fold axes of the capsid. The inserted 7-mer extends into a hydrophobic region near the catalytic center of Car4 to form stable interactions. Mutagenesis studies also identified the key residues in Car4 responsible for the AAV9P31 interaction. These findings provide new insights into the novel receptor recognition mechanism of AAV generated by directed evolution and highlight the application of the BBR method to studying the virus-receptor molecular mechanism. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_38672.map.gz emd_38672.map.gz | 304.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-38672-v30.xml emd-38672-v30.xml emd-38672.xml emd-38672.xml | 12.1 KB 12.1 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_38672.png emd_38672.png | 59.2 KB | ||
| Filedesc metadata |  emd-38672.cif.gz emd-38672.cif.gz | 3.8 KB | ||
| Others |  emd_38672_half_map_1.map.gz emd_38672_half_map_1.map.gz emd_38672_half_map_2.map.gz emd_38672_half_map_2.map.gz | 259.5 MB 259.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-38672 http://ftp.pdbj.org/pub/emdb/structures/EMD-38672 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-38672 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-38672 | HTTPS FTP |
-Validation report
| Summary document |  emd_38672_validation.pdf.gz emd_38672_validation.pdf.gz | 1021.4 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_38672_full_validation.pdf.gz emd_38672_full_validation.pdf.gz | 1021 KB | Display | |
| Data in XML |  emd_38672_validation.xml.gz emd_38672_validation.xml.gz | 16.8 KB | Display | |
| Data in CIF |  emd_38672_validation.cif.gz emd_38672_validation.cif.gz | 19.9 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-38672 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-38672 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-38672 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-38672 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_38672.map.gz / Format: CCP4 / Size: 325 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_38672.map.gz / Format: CCP4 / Size: 325 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.8433 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_38672_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
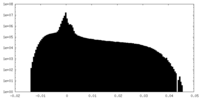
| Projections & Slices |
| ||||||||||||
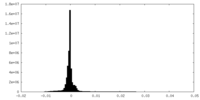
| Density Histograms |
-Half map: #2
| File | emd_38672_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
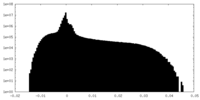
| Projections & Slices |
| ||||||||||||
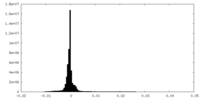
| Density Histograms |
- Sample components
Sample components
-Entire : Adeno-associated virus
| Entire | Name:   Adeno-associated virus Adeno-associated virus |
|---|---|
| Components |
|
-Supramolecule #1: Adeno-associated virus 9
| Supramolecule | Name: Adeno-associated virus 9 / type: virus / ID: 1 / Parent: 0 / NCBI-ID: 235455 / Sci species name: Adeno-associated virus 9 / Sci species strain: 9P31 / Virus type: VIRION / Virus isolate: SEROTYPE / Virus enveloped: No / Virus empty: No |
|---|---|
| Host (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.6 µm / Nominal defocus min: 0.8 µm |
- Image processing
Image processing
| Startup model | Type of model: NONE |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 1.76 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 226745 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)