+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of DDM1-nucleosome complex in ADP state | |||||||||
 Map data Map data | map of DDM1-nucleosome complex in ADP-bound state | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | complex / nucleosome / chromatin remodeling / structural protein-hydrolase-dna complex / GENE REGULATION | |||||||||
| Function / homology |  Function and homology information Function and homology informationDNA-mediated transformation / retrotransposition / chloroplast thylakoid / chromocenter / response to water deprivation / plasmodesma / plant-type vacuole / thylakoid / DNA methylation-dependent constitutive heterochromatin formation / ATP-dependent chromatin remodeler activity ...DNA-mediated transformation / retrotransposition / chloroplast thylakoid / chromocenter / response to water deprivation / plasmodesma / plant-type vacuole / thylakoid / DNA methylation-dependent constitutive heterochromatin formation / ATP-dependent chromatin remodeler activity / plastid / chloroplast stroma / DNA helicase activity / epigenetic regulation of gene expression / chloroplast / response to bacterium / heterochromatin formation / response to wounding / structural constituent of chromatin / nucleosome / peroxisome / DNA helicase / chromatin remodeling / protein heterodimerization activity / nucleolus / ATP hydrolysis activity / DNA binding / extracellular region / ATP binding / nucleus / plasma membrane / cytosol Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.6 Å | |||||||||
 Authors Authors | Liu Y / Zhang Z / Du J | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Plants / Year: 2024 Journal: Nat Plants / Year: 2024Title: Molecular basis of chromatin remodelling by DDM1 involved in plant DNA methylation. Authors: Yue Liu / Zhihui Zhang / Hongmiao Hu / Wei Chen / Fan Zhang / Qian Wang / Changshi Wang / Kaige Yan / Jiamu Du /   Abstract: Eukaryotic gene regulation occurs at the chromatin level, which requires changing the chromatin structure by a group of ATP-dependent DNA translocases-namely, the chromatin remodellers. In plants, ...Eukaryotic gene regulation occurs at the chromatin level, which requires changing the chromatin structure by a group of ATP-dependent DNA translocases-namely, the chromatin remodellers. In plants, chromatin remodellers function in various biological processes and possess both conserved and plant-specific components. DECREASE IN DNA METHYLATION 1 (DDM1) is a plant chromatin remodeller that plays a key role in the maintenance DNA methylation. Here we determined the structures of Arabidopsis DDM1 in complex with nucleosome in ADP-BeF-bound, ADP-bound and nucleotide-free conformations. We show that DDM1 specifically recognizes the H4 tail and nucleosomal DNA. The conformational differences between ADP-BeF-bound, ADP-bound and nucleotide-free DDM1 suggest a chromatin remodelling cycle coupled to ATP binding, hydrolysis and ADP release. This, in turn, triggers conformational changes in the DDM1-bound nucleosomal DNA, which alters the nucleosome structure and promotes DNA sliding. Together, our data reveal the molecular basis of chromatin remodelling by DDM1. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_37533.map.gz emd_37533.map.gz | 5.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-37533-v30.xml emd-37533-v30.xml emd-37533.xml emd-37533.xml | 23.2 KB 23.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_37533_fsc.xml emd_37533_fsc.xml | 8.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_37533.png emd_37533.png | 84 KB | ||
| Filedesc metadata |  emd-37533.cif.gz emd-37533.cif.gz | 7.4 KB | ||
| Others |  emd_37533_half_map_1.map.gz emd_37533_half_map_1.map.gz emd_37533_half_map_2.map.gz emd_37533_half_map_2.map.gz | 40.8 MB 40.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-37533 http://ftp.pdbj.org/pub/emdb/structures/EMD-37533 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-37533 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-37533 | HTTPS FTP |
-Validation report
| Summary document |  emd_37533_validation.pdf.gz emd_37533_validation.pdf.gz | 705.5 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_37533_full_validation.pdf.gz emd_37533_full_validation.pdf.gz | 705.1 KB | Display | |
| Data in XML |  emd_37533_validation.xml.gz emd_37533_validation.xml.gz | 14.8 KB | Display | |
| Data in CIF |  emd_37533_validation.cif.gz emd_37533_validation.cif.gz | 20.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-37533 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-37533 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-37533 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-37533 | HTTPS FTP |
-Related structure data
| Related structure data |  8wh8MC  8wh5C  8wh9C  8whaC  8whbC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_37533.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_37533.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | map of DDM1-nucleosome complex in ADP-bound state | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.08 Å | ||||||||||||||||||||||||||||||||||||
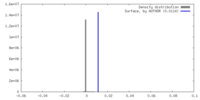
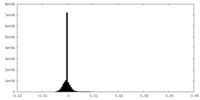
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Half1 map of DDM1-nucleosome complex in ADP-bound state
| File | emd_37533_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half1 map of DDM1-nucleosome complex in ADP-bound state | ||||||||||||
| Projections & Slices |
| ||||||||||||
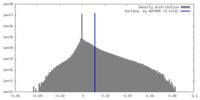
| Density Histograms |
-Half map: Half2 map of DDM1-nucleosome complex in ADP-bound state
| File | emd_37533_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half2 map of DDM1-nucleosome complex in ADP-bound state | ||||||||||||
| Projections & Slices |
| ||||||||||||
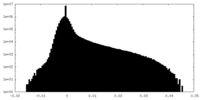
| Density Histograms |
- Sample components
Sample components
-Entire : DDM1-nucleosome complex in ADP state
| Entire | Name: DDM1-nucleosome complex in ADP state |
|---|---|
| Components |
|
-Supramolecule #1: DDM1-nucleosome complex in ADP state
| Supramolecule | Name: DDM1-nucleosome complex in ADP state / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#7 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 290 KDa |
-Macromolecule #1: Histone H3.1
| Macromolecule | Name: Histone H3.1 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 15.300968 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MARTKQTARK STGGKAPRKQ LATKAARKSA PATGGVKKPH RFRPGTVALR EIRKYQKSTE LLIRKLPFQR LVREIAQDFK TDLRFQSSA VAALQEAAEA YLVGLFEDTN LCAIHAKRVT IMPKDIQLAR RIRGERA UniProtKB: Histone H3.1 |
-Macromolecule #2: Histone H4
| Macromolecule | Name: Histone H4 / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 11.436467 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSGRGKGGKG LGKGGAKRHR KVLRDNIQGI TKPAIRRLAR RGGVKRISGL IYEETRGVLK IFLENVIRDA VTYTEHARRK TVTAMDVVY ALKRQGRTLY GFGG UniProtKB: Histone H4 |
-Macromolecule #3: Histone H2A.6
| Macromolecule | Name: Histone H2A.6 / type: protein_or_peptide / ID: 3 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 13.680854 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MAGRGKTLGS GGAKKATSRS SKAGLQFPVG RIARFLKAGK YAERVGAGAP VYLAAVLEYL AAEVLELAGN AARDNKKTRI VPRHIQLAV RNDEELSKLL GDVTIANGGV MPNIHNLLLP KKAGASKPQE D UniProtKB: Histone H2A.6 |
-Macromolecule #4: Histone H2B.6
| Macromolecule | Name: Histone H2B.6 / type: protein_or_peptide / ID: 4 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 16.474459 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MAPRAEKKPA EKKPAAEKPV EEKSKAEKAP AEKKPKAGKK LPKEAGAGGD KKKKMKKKSV ETYKIYIFKV LKQVHPDIGI SSKAMGIMN SFINDIFEKL ASESSKLARY NKKPTITSRE IQTAVRLVLP GELAKHAVSE GTKAVTKFTS S UniProtKB: Histone H2B.6 |
-Macromolecule #7: ATP-dependent DNA helicase DDM1
| Macromolecule | Name: ATP-dependent DNA helicase DDM1 / type: protein_or_peptide / ID: 7 / Number of copies: 1 / Enantiomer: LEVO / EC number: DNA helicase |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 86.844836 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: SMVSLRSRKV IPASEMVSDG KTEKDASGDS PTSVLNEEEN CEEKSVTVVE EEILLAKNGD SSLISEAMAQ EEEQLLKLRE DEEKANNAG SAVAPNLNET QFTKLDELLT QTQLYSEFLL EKMEDITING IESESQKAEP EKTGRGRKRK AASQYNNTKA K RAVAAMIS ...String: SMVSLRSRKV IPASEMVSDG KTEKDASGDS PTSVLNEEEN CEEKSVTVVE EEILLAKNGD SSLISEAMAQ EEEQLLKLRE DEEKANNAG SAVAPNLNET QFTKLDELLT QTQLYSEFLL EKMEDITING IESESQKAEP EKTGRGRKRK AASQYNNTKA K RAVAAMIS RSKEDGETIN SDLTEEETVI KLQNELCPLL TGGQLKSYQL KGVKWLISLW QNGLNGILAD QMGLGKTIQT IG FLSHLKG NGLDGPYLVI APLSTLSNWF NEIARFTPSI NAIIYHGDKN QRDELRRKHM PKTVGPKFPI VITSYEVAMN DAK RILRHY PWKYVVIDEG HRLKNHKCKL LRELKHLKMD NKLLLTGTPL QNNLSELWSL LNFILPDIFT SHDEFESWFD FSEK NKNEA TKEEEEKRRA QVVSKLHGIL RPFILRRMKC DVELSLPRKK EIIMYATMTD HQKKFQEHLV NNTLEAHLGE NAIRG QGWK GKLNNLVIQL RKNCNHPDLL QGQIDGSYLY PPVEEIVGQC GKFRLLERLL VRLFANNHKV LIFSQWTKLL DIMDYY FSE KGFEVCRIDG SVKLDERRRQ IKDFSDEKSS CSIFLLSTRA GGLGINLTAA DTCILYDSDW NPQMDLQAMD RCHRIGQ TK PVHVYRLSTA QSIETRVLKR AYSKLKLEHV VIGQGQFHQE RAKSSTPLEE EDILALLKED ETAEDKLIQT DISDADLD R LLDRSDLTIT APGETQAAEA FPVKGPGWEV VLPSSGGMLS SLNS UniProtKB: ATP-dependent DNA helicase DDM1 |
-Macromolecule #5: DNA (sense strand)
| Macromolecule | Name: DNA (sense strand) / type: dna / ID: 5 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 45.123758 KDa |
| Sequence | String: (DA)(DT)(DC)(DG)(DA)(DG)(DA)(DA)(DT)(DC) (DC)(DC)(DG)(DG)(DT)(DG)(DC)(DC)(DG)(DA) (DG)(DG)(DC)(DC)(DG)(DC)(DT)(DC)(DA) (DA)(DT)(DT)(DG)(DG)(DT)(DC)(DG)(DT)(DA) (DG) (DA)(DC)(DA)(DG)(DC)(DT) ...String: (DA)(DT)(DC)(DG)(DA)(DG)(DA)(DA)(DT)(DC) (DC)(DC)(DG)(DG)(DT)(DG)(DC)(DC)(DG)(DA) (DG)(DG)(DC)(DC)(DG)(DC)(DT)(DC)(DA) (DA)(DT)(DT)(DG)(DG)(DT)(DC)(DG)(DT)(DA) (DG) (DA)(DC)(DA)(DG)(DC)(DT)(DC)(DT) (DA)(DG)(DC)(DA)(DC)(DC)(DG)(DC)(DT)(DT) (DA)(DA) (DA)(DC)(DG)(DC)(DA)(DC)(DG) (DT)(DA)(DC)(DG)(DC)(DG)(DC)(DT)(DG)(DT) (DC)(DC)(DC) (DC)(DC)(DG)(DC)(DG)(DT) (DT)(DT)(DA)(DA)(DC)(DC)(DG)(DC)(DC)(DC) (DA)(DA)(DG)(DG) (DG)(DG)(DA)(DT)(DT) (DA)(DC)(DT)(DC)(DC)(DC)(DT)(DA)(DG)(DT) (DC)(DT)(DC)(DC)(DA) (DG)(DG)(DC)(DA) (DC)(DG)(DT)(DG)(DT)(DC)(DA)(DG)(DA)(DT) (DA)(DT)(DA)(DT)(DA)(DC) (DA)(DT)(DC) (DC)(DG)(DA)(DT) |
-Macromolecule #6: DNA (antisense strand)
| Macromolecule | Name: DNA (antisense strand) / type: dna / ID: 6 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 45.626043 KDa |
| Sequence | String: (DA)(DT)(DC)(DG)(DG)(DA)(DT)(DG)(DT)(DA) (DT)(DA)(DT)(DA)(DT)(DC)(DT)(DG)(DA)(DC) (DA)(DC)(DG)(DT)(DG)(DC)(DC)(DT)(DG) (DG)(DA)(DG)(DA)(DC)(DT)(DA)(DG)(DG)(DG) (DA) (DG)(DT)(DA)(DA)(DT)(DC) ...String: (DA)(DT)(DC)(DG)(DG)(DA)(DT)(DG)(DT)(DA) (DT)(DA)(DT)(DA)(DT)(DC)(DT)(DG)(DA)(DC) (DA)(DC)(DG)(DT)(DG)(DC)(DC)(DT)(DG) (DG)(DA)(DG)(DA)(DC)(DT)(DA)(DG)(DG)(DG) (DA) (DG)(DT)(DA)(DA)(DT)(DC)(DC)(DC) (DC)(DT)(DT)(DG)(DG)(DG)(DC)(DG)(DG)(DT) (DT)(DA) (DA)(DA)(DC)(DG)(DC)(DG)(DG) (DG)(DG)(DG)(DA)(DC)(DA)(DG)(DC)(DG)(DC) (DG)(DT)(DA) (DC)(DG)(DT)(DG)(DC)(DG) (DT)(DT)(DT)(DA)(DA)(DG)(DC)(DG)(DG)(DT) (DG)(DC)(DT)(DA) (DG)(DA)(DG)(DC)(DT) (DG)(DT)(DC)(DT)(DA)(DC)(DG)(DA)(DC)(DC) (DA)(DA)(DT)(DT)(DG) (DA)(DG)(DC)(DG) (DG)(DC)(DC)(DT)(DC)(DG)(DG)(DC)(DA)(DC) (DC)(DG)(DG)(DG)(DA)(DT) (DT)(DC)(DT) (DC)(DG)(DA)(DT) |
-Macromolecule #8: ADENOSINE-5'-DIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-DIPHOSPHATE / type: ligand / ID: 8 / Number of copies: 1 / Formula: ADP |
|---|---|
| Molecular weight | Theoretical: 427.201 Da |
| Chemical component information |  ChemComp-ADP: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2.5 mg/mL |
|---|---|
| Buffer | pH: 8 |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277.15 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Digitization - Frames/image: 1-32 / Number real images: 3025 / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.0 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)