+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
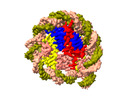
| Title | Structure of nucleosome core particle of Arabidopsis thaliana | |||||||||
 Map data Map data | Arabidopsis nucleosome core particle | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | nucleosome / chromatin / histone / protein dna interaction / nucleoprotein / GENE REGULATION | |||||||||
| Function / homology |  Function and homology information Function and homology informationDNA-mediated transformation / chloroplast thylakoid / chromocenter / thylakoid / response to water deprivation / plasmodesma / plant-type vacuole / chloroplast stroma / plastid / chloroplast ...DNA-mediated transformation / chloroplast thylakoid / chromocenter / thylakoid / response to water deprivation / plasmodesma / plant-type vacuole / chloroplast stroma / plastid / chloroplast / response to bacterium / response to wounding / structural constituent of chromatin / peroxisome / nucleosome / protein heterodimerization activity / nucleolus / DNA binding / extracellular region / nucleus / plasma membrane / cytosol Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.17 Å | |||||||||
 Authors Authors | Liu Y / Zhang Z / Du J | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Plants / Year: 2024 Journal: Nat Plants / Year: 2024Title: Molecular basis of chromatin remodelling by DDM1 involved in plant DNA methylation. Authors: Yue Liu / Zhihui Zhang / Hongmiao Hu / Wei Chen / Fan Zhang / Qian Wang / Changshi Wang / Kaige Yan / Jiamu Du /   Abstract: Eukaryotic gene regulation occurs at the chromatin level, which requires changing the chromatin structure by a group of ATP-dependent DNA translocases-namely, the chromatin remodellers. In plants, ...Eukaryotic gene regulation occurs at the chromatin level, which requires changing the chromatin structure by a group of ATP-dependent DNA translocases-namely, the chromatin remodellers. In plants, chromatin remodellers function in various biological processes and possess both conserved and plant-specific components. DECREASE IN DNA METHYLATION 1 (DDM1) is a plant chromatin remodeller that plays a key role in the maintenance DNA methylation. Here we determined the structures of Arabidopsis DDM1 in complex with nucleosome in ADP-BeF-bound, ADP-bound and nucleotide-free conformations. We show that DDM1 specifically recognizes the H4 tail and nucleosomal DNA. The conformational differences between ADP-BeF-bound, ADP-bound and nucleotide-free DDM1 suggest a chromatin remodelling cycle coupled to ATP binding, hydrolysis and ADP release. This, in turn, triggers conformational changes in the DDM1-bound nucleosomal DNA, which alters the nucleosome structure and promotes DNA sliding. Together, our data reveal the molecular basis of chromatin remodelling by DDM1. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_37538.map.gz emd_37538.map.gz | 5.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-37538-v30.xml emd-37538-v30.xml emd-37538.xml emd-37538.xml | 23.7 KB 23.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_37538_fsc.xml emd_37538_fsc.xml | 9.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_37538.png emd_37538.png | 99 KB | ||
| Filedesc metadata |  emd-37538.cif.gz emd-37538.cif.gz | 6.9 KB | ||
| Others |  emd_37538_half_map_1.map.gz emd_37538_half_map_1.map.gz emd_37538_half_map_2.map.gz emd_37538_half_map_2.map.gz | 49.4 MB 49.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-37538 http://ftp.pdbj.org/pub/emdb/structures/EMD-37538 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-37538 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-37538 | HTTPS FTP |
-Related structure data
| Related structure data |  8whbMC  8wh5C  8wh8C  8wh9C  8whaC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_37538.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_37538.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Arabidopsis nucleosome core particle | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.855 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Half1 map of Arabidopsis nucleosome core particle
| File | emd_37538_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half1 map of Arabidopsis nucleosome core particle | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half2 map of Arabidopsis nucleosome core particle
| File | emd_37538_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half2 map of Arabidopsis nucleosome core particle | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Structure of the nucleosome core particle of Arabidopsis thaliana
| Entire | Name: Structure of the nucleosome core particle of Arabidopsis thaliana |
|---|---|
| Components |
|
-Supramolecule #1: Structure of the nucleosome core particle of Arabidopsis thaliana
| Supramolecule | Name: Structure of the nucleosome core particle of Arabidopsis thaliana type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 200 KDa |
-Macromolecule #1: Histone H3.1
| Macromolecule | Name: Histone H3.1 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 15.300968 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MARTKQTARK STGGKAPRKQ LATKAARKSA PATGGVKKPH RFRPGTVALR EIRKYQKSTE LLIRKLPFQR LVREIAQDFK TDLRFQSSA VAALQEAAEA YLVGLFEDTN LCAIHAKRVT IMPKDIQLAR RIRGERA UniProtKB: Histone H3.1 |
-Macromolecule #2: Histone H4
| Macromolecule | Name: Histone H4 / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 11.436467 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSGRGKGGKG LGKGGAKRHR KVLRDNIQGI TKPAIRRLAR RGGVKRISGL IYEETRGVLK IFLENVIRDA VTYTEHARRK TVTAMDVVY ALKRQGRTLY GFGG UniProtKB: Histone H4 |
-Macromolecule #3: Histone H2A.6
| Macromolecule | Name: Histone H2A.6 / type: protein_or_peptide / ID: 3 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 13.680854 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MAGRGKTLGS GGAKKATSRS SKAGLQFPVG RIARFLKAGK YAERVGAGAP VYLAAVLEYL AAEVLELAGN AARDNKKTRI VPRHIQLAV RNDEELSKLL GDVTIANGGV MPNIHNLLLP KKAGASKPQE D UniProtKB: Histone H2A.6 |
-Macromolecule #4: Histone H2B.6
| Macromolecule | Name: Histone H2B.6 / type: protein_or_peptide / ID: 4 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 16.474459 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MAPRAEKKPA EKKPAAEKPV EEKSKAEKAP AEKKPKAGKK LPKEAGAGGD KKKKMKKKSV ETYKIYIFKV LKQVHPDIGI SSKAMGIMN SFINDIFEKL ASESSKLARY NKKPTITSRE IQTAVRLVLP GELAKHAVSE GTKAVTKFTS S UniProtKB: Histone H2B.6 |
-Macromolecule #5: DNA (sense strand)
| Macromolecule | Name: DNA (sense strand) / type: dna / ID: 5 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 45.123758 KDa |
| Sequence | String: (DA)(DT)(DC)(DG)(DA)(DG)(DA)(DA)(DT)(DC) (DC)(DC)(DG)(DG)(DT)(DG)(DC)(DC)(DG)(DA) (DG)(DG)(DC)(DC)(DG)(DC)(DT)(DC)(DA) (DA)(DT)(DT)(DG)(DG)(DT)(DC)(DG)(DT)(DA) (DG) (DA)(DC)(DA)(DG)(DC)(DT) ...String: (DA)(DT)(DC)(DG)(DA)(DG)(DA)(DA)(DT)(DC) (DC)(DC)(DG)(DG)(DT)(DG)(DC)(DC)(DG)(DA) (DG)(DG)(DC)(DC)(DG)(DC)(DT)(DC)(DA) (DA)(DT)(DT)(DG)(DG)(DT)(DC)(DG)(DT)(DA) (DG) (DA)(DC)(DA)(DG)(DC)(DT)(DC)(DT) (DA)(DG)(DC)(DA)(DC)(DC)(DG)(DC)(DT)(DT) (DA)(DA) (DA)(DC)(DG)(DC)(DA)(DC)(DG) (DT)(DA)(DC)(DG)(DC)(DG)(DC)(DT)(DG)(DT) (DC)(DC)(DC) (DC)(DC)(DG)(DC)(DG)(DT) (DT)(DT)(DA)(DA)(DC)(DC)(DG)(DC)(DC)(DC) (DA)(DA)(DG)(DG) (DG)(DG)(DA)(DT)(DT) (DA)(DC)(DT)(DC)(DC)(DC)(DT)(DA)(DG)(DT) (DC)(DT)(DC)(DC)(DA) (DG)(DG)(DC)(DA) (DC)(DG)(DT)(DG)(DT)(DC)(DA)(DG)(DA)(DT) (DA)(DT)(DA)(DT)(DA)(DC) (DA)(DT)(DC) (DC)(DG)(DA)(DT) |
-Macromolecule #6: DNA (antisense strand)
| Macromolecule | Name: DNA (antisense strand) / type: dna / ID: 6 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 45.626043 KDa |
| Sequence | String: (DA)(DT)(DC)(DG)(DG)(DA)(DT)(DG)(DT)(DA) (DT)(DA)(DT)(DA)(DT)(DC)(DT)(DG)(DA)(DC) (DA)(DC)(DG)(DT)(DG)(DC)(DC)(DT)(DG) (DG)(DA)(DG)(DA)(DC)(DT)(DA)(DG)(DG)(DG) (DA) (DG)(DT)(DA)(DA)(DT)(DC) ...String: (DA)(DT)(DC)(DG)(DG)(DA)(DT)(DG)(DT)(DA) (DT)(DA)(DT)(DA)(DT)(DC)(DT)(DG)(DA)(DC) (DA)(DC)(DG)(DT)(DG)(DC)(DC)(DT)(DG) (DG)(DA)(DG)(DA)(DC)(DT)(DA)(DG)(DG)(DG) (DA) (DG)(DT)(DA)(DA)(DT)(DC)(DC)(DC) (DC)(DT)(DT)(DG)(DG)(DG)(DC)(DG)(DG)(DT) (DT)(DA) (DA)(DA)(DC)(DG)(DC)(DG)(DG) (DG)(DG)(DG)(DA)(DC)(DA)(DG)(DC)(DG)(DC) (DG)(DT)(DA) (DC)(DG)(DT)(DG)(DC)(DG) (DT)(DT)(DT)(DA)(DA)(DG)(DC)(DG)(DG)(DT) (DG)(DC)(DT)(DA) (DG)(DA)(DG)(DC)(DT) (DG)(DT)(DC)(DT)(DA)(DC)(DG)(DA)(DC)(DC) (DA)(DA)(DT)(DT)(DG) (DA)(DG)(DC)(DG) (DG)(DC)(DC)(DT)(DC)(DG)(DG)(DC)(DA)(DC) (DC)(DG)(DG)(DG)(DA)(DT) (DT)(DC)(DT) (DC)(DG)(DA)(DT) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2.5 mg/mL |
|---|---|
| Buffer | pH: 8 |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277.15 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Detector mode: SUPER-RESOLUTION / Digitization - Frames/image: 1-32 / Number real images: 6776 / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.0 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)