+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of PAO1-ImcA with GMPCPP | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | GGDEF domain / diguanylate cyclase activity / membrane protein | |||||||||
| Function / homology |  Function and homology information Function and homology information | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.2 Å | |||||||||
 Authors Authors | Zhan XL / Zhang K / Wang CC / Fan Q / Tang XJ / Zhang X / Wang K / Fu Y / Liang HH | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2024 Journal: Nat Commun / Year: 2024Title: A c-di-GMP signaling module controls responses to iron in Pseudomonas aeruginosa. Authors: Xueliang Zhan / Kuo Zhang / Chenchen Wang / Qiao Fan / Xiujia Tang / Xi Zhang / Ke Wang / Yang Fu / Haihua Liang /  Abstract: Cyclic dimeric guanosine monophosphate (c-di-GMP) serves as a bacterial second messenger that modulates various processes including biofilm formation, motility, and host-microbe symbiosis. Numerous ...Cyclic dimeric guanosine monophosphate (c-di-GMP) serves as a bacterial second messenger that modulates various processes including biofilm formation, motility, and host-microbe symbiosis. Numerous studies have conducted comprehensive analysis of c-di-GMP. However, the mechanisms by which certain environmental signals such as iron control intracellular c-di-GMP levels are unclear. Here, we show that iron regulates c-di-GMP levels in Pseudomonas aeruginosa by modulating the interaction between an iron-sensing protein, IsmP, and a diguanylate cyclase, ImcA. Binding of iron to the CHASE4 domain of IsmP inhibits the IsmP-ImcA interaction, which leads to increased c-di-GMP synthesis by ImcA, thus promoting biofilm formation and reducing bacterial motility. Structural characterization of the apo-CHASE4 domain and its binding to iron allows us to pinpoint residues defining its specificity. In addition, the cryo-electron microscopy structure of ImcA in complex with a c-di-GMP analog (GMPCPP) suggests a unique conformation in which the compound binds to the catalytic pockets and to the membrane-proximal side located at the cytoplasm. Thus, our results indicate that a CHASE4 domain directly senses iron and modulates the crosstalk between c-di-GMP metabolic enzymes. | |||||||||
| History |
|
- Structure visualization
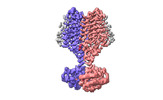
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_37444.map.gz emd_37444.map.gz | 165.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-37444-v30.xml emd-37444-v30.xml emd-37444.xml emd-37444.xml | 14.1 KB 14.1 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_37444.png emd_37444.png | 53.2 KB | ||
| Filedesc metadata |  emd-37444.cif.gz emd-37444.cif.gz | 5.5 KB | ||
| Others |  emd_37444_half_map_1.map.gz emd_37444_half_map_1.map.gz emd_37444_half_map_2.map.gz emd_37444_half_map_2.map.gz | 164.9 MB 164.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-37444 http://ftp.pdbj.org/pub/emdb/structures/EMD-37444 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-37444 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-37444 | HTTPS FTP |
-Validation report
| Summary document |  emd_37444_validation.pdf.gz emd_37444_validation.pdf.gz | 756.3 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_37444_full_validation.pdf.gz emd_37444_full_validation.pdf.gz | 755.9 KB | Display | |
| Data in XML |  emd_37444_validation.xml.gz emd_37444_validation.xml.gz | 15 KB | Display | |
| Data in CIF |  emd_37444_validation.cif.gz emd_37444_validation.cif.gz | 17.7 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-37444 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-37444 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-37444 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-37444 | HTTPS FTP |
-Related structure data
| Related structure data |  8wcnMC  8wctC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_37444.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_37444.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.668 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_37444_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
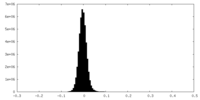
| Density Histograms |
-Half map: #1
| File | emd_37444_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
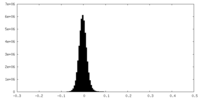
| Density Histograms |
- Sample components
Sample components
-Entire : GGDEF domain-containing protein
| Entire | Name: GGDEF domain-containing protein |
|---|---|
| Components |
|
-Supramolecule #1: GGDEF domain-containing protein
| Supramolecule | Name: GGDEF domain-containing protein / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Diguanylate cyclase
| Macromolecule | Name: Diguanylate cyclase / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 51.329805 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGSDYDIPTT ENLYFQGSML ARDSLVQAGL PDNPYARQLR NGFRWLRFEK ELENEFREFL SWNSLMQRRA AIGVAFLIWA LFIVADWMM VDIRLHPSLF EQLLGVRLGM IGLLLVVWPA AFLPSLRKVG DAIAPYCLLL INLAVLACDV LFEWHGVPRF T QLGATLGI ...String: MGSDYDIPTT ENLYFQGSML ARDSLVQAGL PDNPYARQLR NGFRWLRFEK ELENEFREFL SWNSLMQRRA AIGVAFLIWA LFIVADWMM VDIRLHPSLF EQLLGVRLGM IGLLLVVWPA AFLPSLRKVG DAIAPYCLLL INLAVLACDV LFEWHGVPRF T QLGATLGI LAVFFPLGLA FWACVRLALL CLALNLAVFL LFGGEENLRT NLLNTLYNGL VVLICSFALY LQDYAQREQF LG RRLLGMM AEQDSLTGLV NRRYYELLAQ RALEQGAREE KGVALILVDV DDFKAYNDHY GHPAGDAALR QLGVVLRQGA RRP LDIAAR LGGEEFAVLL YDSEEGNTLA IAERLRQAVE ALGIEHLGSS AGPCLTISLG VAYSTSGMGL DALYREADRA LYEA KDAGR NAVRVAFRQH DRLEGSFLSA WSHPQFEKGG GSGGGSGGGS WSHPQFEKLE HHHHHH UniProtKB: diguanylate cyclase |
-Macromolecule #2: PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER
| Macromolecule | Name: PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER / type: ligand / ID: 2 / Number of copies: 4 / Formula: G2P |
|---|---|
| Molecular weight | Theoretical: 521.208 Da |
| Chemical component information |  ChemComp-G2P: |
-Macromolecule #3: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 3 / Number of copies: 2 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 58.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.7 µm / Nominal defocus min: 0.8 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: OTHER |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.2 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 205294 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)