[English] 日本語
 Yorodumi
Yorodumi- EMDB-37266: Cryo-EM structure of the KLHL22 E3 ligase bound to human glutamat... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the KLHL22 E3 ligase bound to human glutamate dehydrogenase I | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | E3 ligase / STRUCTURAL PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationL-leucine binding / glutamate dehydrogenase [NAD(P)+] activity / glutamate catabolic process / tricarboxylic acid metabolic process / glutamate dehydrogenase [NAD(P)+] / glutamate biosynthetic process / polar microtubule / glutamate dehydrogenase (NADP+) activity / Glutamate and glutamine metabolism / glutamate dehydrogenase (NAD+) activity ...L-leucine binding / glutamate dehydrogenase [NAD(P)+] activity / glutamate catabolic process / tricarboxylic acid metabolic process / glutamate dehydrogenase [NAD(P)+] / glutamate biosynthetic process / polar microtubule / glutamate dehydrogenase (NADP+) activity / Glutamate and glutamine metabolism / glutamate dehydrogenase (NAD+) activity / positive regulation of T cell mediated immune response to tumor cell / cellular response to L-leucine / negative regulation of type I interferon production / Cul3-RING ubiquitin ligase complex / intercellular bridge / glutamine metabolic process / mitotic spindle assembly checkpoint signaling / protein monoubiquitination / mitotic sister chromatid segregation / NAD+ binding / ubiquitin-like ligase-substrate adaptor activity / 14-3-3 protein binding / Mitochondrial protein degradation / substantia nigra development / positive regulation of TORC1 signaling / negative regulation of autophagy / cellular response to amino acid stimulus / ADP binding / Transcriptional activation of mitochondrial biogenesis / positive regulation of insulin secretion / mitotic spindle / microtubule cytoskeleton / positive regulation of T cell activation / Antigen processing: Ubiquitination & Proteasome degradation / Neddylation / ubiquitin-dependent protein catabolic process / positive regulation of cell growth / proteasome-mediated ubiquitin-dependent protein catabolic process / lysosome / mitochondrial matrix / cell division / intracellular membrane-bounded organelle / centrosome / GTP binding / endoplasmic reticulum / protein homodimerization activity / mitochondrion / ATP binding / nucleus / cytosol / cytoplasm Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.06 Å | |||||||||
 Authors Authors | Su M-Y | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Structure / Year: 2023 Journal: Structure / Year: 2023Title: Cryo-EM structure of the KLHL22 E3 ligase bound to an oligomeric metabolic enzyme. Authors: Fei Teng / Yang Wang / Ming Liu / Shuyun Tian / Goran Stjepanovic / Ming-Yuan Su /  Abstract: CULLIN-RING ligases constitute the largest group of E3 ubiquitin ligases. While some CULLIN family members recruit adapters before engaging further with different substrate receptors, homo-dimeric ...CULLIN-RING ligases constitute the largest group of E3 ubiquitin ligases. While some CULLIN family members recruit adapters before engaging further with different substrate receptors, homo-dimeric BTB-Kelch family proteins combine adapter and substrate receptor into a single polypeptide for the CULLIN3 family. However, the entire structural assembly and molecular details have not been elucidated to date. Here, we present a cryo-EM structure of the CULLIN3 in complex with Kelch-like protein 22 (KLHL22) and a mitochondrial glutamate dehydrogenase complex I (GDH1) at 3.06 Å resolution. The structure adopts a W-shaped architecture formed by E3 ligase dimers. Three CULLIN3 dimers were found to be dynamically associated with a single GDH1 hexamer. CULLIN3 ligase mediated the polyubiquitination of GDH1 in vitro. Together, these results enabled the establishment of a structural model for understanding the complete assembly of BTB-Kelch proteins with CULLIN3 and how together they recognize oligomeric substrates and target them for ubiquitination. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_37266.map.gz emd_37266.map.gz | 211.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-37266-v30.xml emd-37266-v30.xml emd-37266.xml emd-37266.xml | 17.2 KB 17.2 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_37266.png emd_37266.png | 34.4 KB | ||
| Filedesc metadata |  emd-37266.cif.gz emd-37266.cif.gz | 6.1 KB | ||
| Others |  emd_37266_additional_1.map.gz emd_37266_additional_1.map.gz emd_37266_half_map_1.map.gz emd_37266_half_map_1.map.gz emd_37266_half_map_2.map.gz emd_37266_half_map_2.map.gz | 768.7 MB 391.4 MB 391.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-37266 http://ftp.pdbj.org/pub/emdb/structures/EMD-37266 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-37266 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-37266 | HTTPS FTP |
-Validation report
| Summary document |  emd_37266_validation.pdf.gz emd_37266_validation.pdf.gz | 943.4 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_37266_full_validation.pdf.gz emd_37266_full_validation.pdf.gz | 943 KB | Display | |
| Data in XML |  emd_37266_validation.xml.gz emd_37266_validation.xml.gz | 18.2 KB | Display | |
| Data in CIF |  emd_37266_validation.cif.gz emd_37266_validation.cif.gz | 21.5 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-37266 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-37266 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-37266 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-37266 | HTTPS FTP |
-Related structure data
| Related structure data |  8w4jMC  8kgyC  8khpC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_37266.map.gz / Format: CCP4 / Size: 421.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_37266.map.gz / Format: CCP4 / Size: 421.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.85 Å | ||||||||||||||||||||||||||||||||||||
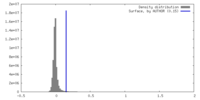
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: #1
| File | emd_37266_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
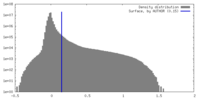
| Density Histograms |
-Half map: #2
| File | emd_37266_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
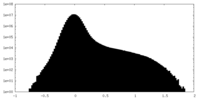
| Density Histograms |
-Half map: #1
| File | emd_37266_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : the CULLIN3-KLHL22-RBX1 E3 ligase bound to glutamate dehydrogenase I
| Entire | Name: the CULLIN3-KLHL22-RBX1 E3 ligase bound to glutamate dehydrogenase I |
|---|---|
| Components |
|
-Supramolecule #1: the CULLIN3-KLHL22-RBX1 E3 ligase bound to glutamate dehydrogenase I
| Supramolecule | Name: the CULLIN3-KLHL22-RBX1 E3 ligase bound to glutamate dehydrogenase I type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Glutamate dehydrogenase 1, mitochondrial
| Macromolecule | Name: Glutamate dehydrogenase 1, mitochondrial / type: protein_or_peptide / ID: 1 / Number of copies: 6 / Enantiomer: LEVO / EC number: glutamate dehydrogenase [NAD(P)+] |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 61.480746 KDa |
| Sequence | String: MYRYLGEALL LSRAGPAALG SASADSAALL GWARGQPAAA PQPGLALAAR RHYSEAVADR EDDPNFFKMV EGFFDRGASI VEDKLVEDL RTRESEEQKR NRVRGILRII KPCNHVLSLS FPIRRDDGSW EVIEGYRAQH SQHRTPCKGG IRYSTDVSVD E VKALASLM ...String: MYRYLGEALL LSRAGPAALG SASADSAALL GWARGQPAAA PQPGLALAAR RHYSEAVADR EDDPNFFKMV EGFFDRGASI VEDKLVEDL RTRESEEQKR NRVRGILRII KPCNHVLSLS FPIRRDDGSW EVIEGYRAQH SQHRTPCKGG IRYSTDVSVD E VKALASLM TYKCAVVDVP FGGAKAGVKI NPKNYTDNEL EKITRRFTME LAKKGFIGPG IDVPAPDMST GEREMSWIAD TY ASTIGHY DINAHACVTG KPISQGGIHG RISATGRGVF HGIENFINEA SYMSILGMTP GFGDKTFVVQ GFGNVGLHSM RYL HRFGAK CIAVGESDGS IWNPDGIDPK ELEDFKLQHG SILGFPKAKP YEGSILEADC DILIPAASEK QLTKSNAPRV KAKI IAEGA NGPTTPEADK IFLERNIMVI PDLYLNAGGV TVSYFEWLKN LNHVSYGRLT FKYERDSNYH LLMSVQESLE RKFGK HGGT IPIVPTAEFQ DRISGASEKD IVHSGLAYTM ERSARQIMRT AMKYNLGLDL RTAAYVNAIE KVFKVYNEAG VTFT UniProtKB: Glutamate dehydrogenase 1, mitochondrial |
-Macromolecule #2: Kelch-like protein 22
| Macromolecule | Name: Kelch-like protein 22 / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 71.744594 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MAEEQEFTQL CKLPAQPSHP HCVNNTYRSA QHSQALLRGL LALRDSGILF DVVLVVEGRH IEAHRILLAA SCDYFRGMFA GGLKEMEQE EVLIHGVSYN AMCQILHFIY TSELELSLSN VQETLVAACQ LQIPEIIHFC CDFLMSWVDE ENILDVYRLA E LFDLSRLT ...String: MAEEQEFTQL CKLPAQPSHP HCVNNTYRSA QHSQALLRGL LALRDSGILF DVVLVVEGRH IEAHRILLAA SCDYFRGMFA GGLKEMEQE EVLIHGVSYN AMCQILHFIY TSELELSLSN VQETLVAACQ LQIPEIIHFC CDFLMSWVDE ENILDVYRLA E LFDLSRLT EQLDTYILKN FVAFSRTDKY RQLPLEKVYS LLSSNRLEVS CETEVYEGAL LYHYSLEQVQ ADQISLHEPP KL LETVRFP LMEAEVLQRL HDKLDPSPLR DTVASALMYH RNESLQPSLQ SPQTELRSDF QCVVGFGGIH STPSTVLSDQ AKY LNPLLG EWKHFTASLA PRMSNQGIAV LNNFVYLIGG DNNVQGFRAE SRCWRYDPRH NRWFQIQSLQ QEHADLSVCV VGRY IYAVA GRDYHNDLNA VERYDPATNS WAYVAPLKRE VYAHAGATLE GKMYITCGRR GEDYLKETHC YDPGSNTWHT LADGP VRRA WHGMATLLNK LYVIGGSNND AGYRRDVHQV ACYSCTSGQW SSVCPLPAGH GEPGIAVLDN RIYVLGGRSH NRGSRT GYV HIYDVEKDCW EEGPQLDNSI SGLAACVLTL PRSLLLEPPR GTPDRSQADP DFASEVMSVS DWEEFDNSSE D UniProtKB: Kelch-like protein 22 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 1.072 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.9000000000000001 µm / Nominal defocus min: 1.1 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: OTHER |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.06 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 62817 |
| Initial angle assignment | Type: OTHER |
| Final angle assignment | Type: OTHER |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)