+ Open data
Open data
 Loading...
Loading...
- Basic information
Basic information
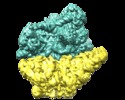
| Entry | Database: EMDB / ID: EMD-3551 | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of the yeast mitochondrial ribosome - Class A | ||||||||||||||||||
 Map data Map data | |||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||
 Keywords Keywords | yeast / mitochondrial / ribosome / saccharomyces cerevisiae | ||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationBranched-chain amino acid catabolism / 3-hydroxyisobutyryl-CoA hydrolase / positive regulation of mitochondrial DNA replication / 3-hydroxyisobutyryl-CoA hydrolase activity / mitochondrial translational initiation / L-valine catabolic process / : / mitochondrial respiratory chain complex IV assembly / Mitochondrial protein degradation / ribonuclease III activity ...Branched-chain amino acid catabolism / 3-hydroxyisobutyryl-CoA hydrolase / positive regulation of mitochondrial DNA replication / 3-hydroxyisobutyryl-CoA hydrolase activity / mitochondrial translational initiation / L-valine catabolic process / : / mitochondrial respiratory chain complex IV assembly / Mitochondrial protein degradation / ribonuclease III activity / mitochondrial ribosome assembly / DNA strand exchange activity / mitochondrial large ribosomal subunit / mitochondrial ribosome / mitochondrial small ribosomal subunit / mitochondrial translation / sporulation resulting in formation of a cellular spore / superoxide dismutase activity / RNA processing / cell redox homeostasis / mRNA processing / peroxisome / single-stranded DNA binding / double-stranded RNA binding / large ribosomal subunit / ribosome biogenesis / transferase activity / ribosomal small subunit biogenesis / ribosomal small subunit assembly / cellular response to oxidative stress / small ribosomal subunit / small ribosomal subunit rRNA binding / large ribosomal subunit rRNA binding / DNA recombination / cytosolic large ribosomal subunit / mitochondrial inner membrane / negative regulation of translation / rRNA binding / structural constituent of ribosome / ribosome / translation / mRNA binding / regulation of DNA-templated transcription / GTP binding / mitochondrion / DNA binding / RNA binding / zinc ion binding / ATP binding / metal ion binding / nucleus / cytoplasm Similarity search - Function 54S ribosomal protein L8, C-terminal / Ribosomal L27 protein, C-terminal / Ribosomal L27 protein C-terminal domain / 54S ribosomal protein L8 C-terminal domain / Large ribosomal subunit protein uL24m / Ribosomal protein VAR1 / Mitochondrial ribosomal protein (VAR1) / Ribosomal protein L31, mitochondrial / Ribosomal protein L20, mitochondrial / Mitochondrial homologous recombination protein 1 ...54S ribosomal protein L8, C-terminal / Ribosomal L27 protein, C-terminal / Ribosomal L27 protein C-terminal domain / 54S ribosomal protein L8 C-terminal domain / Large ribosomal subunit protein uL24m / Ribosomal protein VAR1 / Mitochondrial ribosomal protein (VAR1) / Ribosomal protein L31, mitochondrial / Ribosomal protein L20, mitochondrial / Mitochondrial homologous recombination protein 1 / 54S ribosomal protein L25 / 54S ribosomal protein L15, mitochondrial / MRPL25 domain / 54S ribosomal protein L3, double-stranded RNA binding domain / Mitochondrial ribosomal protein subunit L20 / Transcriptional regulation of mitochondrial recombination / Mitochondrial ribosomal protein mL59 / Mitochondrial ribosomal protein L31 / 54S ribosomal protein L36, yeast / 54S ribosomal protein L28, mitochondrial / : / : / : / Ribosomal protein L31p, N-terminal / Mitochondrial ribosomal protein MRP51, fungi / Mitochondrial ribosomal protein S25 / Ribosomal protein S24, mitochondrial / Ribosomal protein S23, mitochondrial, fungi / Mitochondrial ribosomal protein subunit / Mitochondrial ribosomal protein S25 / Ribosomal protein S35, mitochondrial / Enoyl-CoA hydratase/isomerase, HIBYL-CoA-H type / Ribosomal protein MRP10, mitochondrial / Eukaryotic mitochondrial regulator protein / Enoyl-CoA hydratase/isomerase domain / Enoyl-CoA hydratase/isomerase / Protein Fyv4 / Small ribosomal subunit protein mS41 SAM domain / IGR protein motif / IGR / Ribonuclease-III-like / Ribonuclease III domain / Ribonuclease III family domain profile. / Ribonuclease III family / Ribonuclease III domain / Enoyl-CoA hydratase/isomerase, conserved site / Enoyl-CoA hydratase/isomerase signature. / : / Ribosomal protein S27/S33, mitochondrial / Ribosomal protein S24/S35, mitochondrial / Mitochondrial ribosomal subunit S27 / Ribosomal protein S24/S35, mitochondrial, conserved domain / Mitochondrial ribosomal subunit protein / Ribosomal protein L28/L40, mitochondrial / Mitochondrial ribosomal protein L28 / : / Ribosomal protein S23/S29, mitochondrial / Mitochondrial ribosomal death-associated protein 3 / Mitochondrial ribosomal protein L46 NUDIX / Ribosomal protein L50, mitochondria / Manganese/iron superoxide dismutase, C-terminal / Manganese/iron superoxide dismutase, C-terminal domain superfamily / Manganese/iron superoxide dismutase, N-terminal domain superfamily / Iron/manganese superoxide dismutases, C-terminal domain / Ribosomal subunit 39S / Ribosomal protein L46, N-terminal / 39S mitochondrial ribosomal protein L46 / Ribosomal protein L49/IMG2 / Mitochondrial large subunit ribosomal protein (Img2) / Mitochondrial mRNA-processing protein COX24, C-terminal / Ribosomal protein L27/L41, mitochondrial / : / Mitochondrial mRNA-processing protein COX24, C-terminal / Mitochondrial ribosomal protein L27 / Mitochondrial domain of unknown function (DUF1713) / 39S ribosomal protein L46, mitochondrial / Ribosomal protein L47, mitochondrial / MRP-L47 superfamily, mitochondrial / 39S ribosomal protein L43/54S ribosomal protein L51 / Mitochondrial 39-S ribosomal protein L47 (MRP-L47) / Phosphatidylethanolamine-binding protein, eukaryotic / Phosphatidylethanolamine-binding protein / Phosphatidylethanolamine-binding protein / PEBP-like superfamily / Double-stranded RNA binding motif / Double-stranded RNA binding motif / Ribonuclease III, endonuclease domain superfamily / Double stranded RNA-binding domain (dsRBD) profile. / Double-stranded RNA-binding domain / CHCH / CHCH domain / Coiled coil-helix-coiled coil-helix (CHCH) domain profile. / Mitochondrial ribosomal protein L51 / S25 / CI-B8 domain / Ribosomal protein/NADH dehydrogenase domain / Mitochondrial ribosomal protein L51 / S25 / CI-B8 domain / ClpP/crotonase-like domain superfamily / Ribosomal protein S16, conserved site / Ribosomal protein S16 signature. / Ribosomal protein S21 / Ribosomal protein S21 Similarity search - Domain/homology Large ribosomal subunit protein bL36m / Small ribosomal subunit protein mS37 / Small ribosomal subunit protein uS3m / Small ribosomal subunit protein mS43 / Small ribosomal subunit protein uS14m / Small ribosomal subunit protein mS27 / Large ribosomal subunit protein bL27m / Large ribosomal subunit protein mL60 / Small ribosomal subunit protein mS26 / Large ribosomal subunit protein uL30m ...Large ribosomal subunit protein bL36m / Small ribosomal subunit protein mS37 / Small ribosomal subunit protein uS3m / Small ribosomal subunit protein mS43 / Small ribosomal subunit protein uS14m / Small ribosomal subunit protein mS27 / Large ribosomal subunit protein bL27m / Large ribosomal subunit protein mL60 / Small ribosomal subunit protein mS26 / Large ribosomal subunit protein uL30m / Small ribosomal subunit protein uS15m / Large ribosomal subunit protein bL17m / Large ribosomal subunit protein mL58 / Large ribosomal subunit protein mL59 / Large ribosomal subunit protein bL32m / Large ribosomal subunit protein bL19m / Large ribosomal subunit protein mL49 / Small ribosomal subunit protein uS4m / Small ribosomal subunit protein bS6m / Small ribosomal subunit protein mS47 / Large ribosomal subunit protein uL3m / Small ribosomal subunit protein mS38 / Large ribosomal subunit protein uL23m / Large ribosomal subunit protein uL2m / Small ribosomal subunit protein uS2m / Large ribosomal subunit protein uL6m / Small ribosomal subunit protein uS5m / Large ribosomal subunit protein uL14m / Large ribosomal subunit protein mL44 / Large ribosomal subunit protein uL29m / Large ribosomal subunit protein uL5m / Large ribosomal subunit protein uL15m / Large ribosomal subunit protein mL57 / Large ribosomal subunit protein bL28m / Large ribosomal subunit protein mL41 / Large ribosomal subunit protein mL40 / Large ribosomal subunit protein mL46 / Large ribosomal subunit protein bL31m / Large ribosomal subunit protein bL33m / Large ribosomal subunit protein uL24m / Large ribosomal subunit protein uL16m / Small ribosomal subunit protein uS9m / Small ribosomal subunit protein bS21m / Small ribosomal subunit protein mS41 / Small ribosomal subunit protein bS18m / Small ribosomal subunit protein mS23 / Large ribosomal subunit protein bL21m / Small ribosomal subunit protein uS11m / Small ribosomal subunit protein mS42 / Small ribosomal subunit protein uS7m / Large ribosomal subunit protein uL4m / Small ribosomal subunit protein mS45 / Small ribosomal subunit protein mS33 / Large ribosomal subunit protein bL9m / Small ribosomal subunit protein uS12m / Small ribosomal subunit protein uS19m / Large ribosomal subunit protein uL22m / Large ribosomal subunit protein bL35m / Small ribosomal subunit protein uS13m / Small ribosomal subunit protein mS29 / Large ribosomal subunit protein mL50 / Small ribosomal subunit protein bS16m / Small ribosomal subunit protein bS1m / Small ribosomal subunit protein uS10m / Small ribosomal subunit protein uS17m / Small ribosomal subunit protein mS46 / Small ribosomal subunit protein uS8m / Small ribosomal subunit protein mS35 / Large ribosomal subunit protein bL34m / Large ribosomal subunit protein mL43 / Large ribosomal subunit protein mL67 / Large ribosomal subunit protein mL38 / Large ribosomal subunit protein uL13m Similarity search - Component | ||||||||||||||||||
| Biological species |  | ||||||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.25 Å | ||||||||||||||||||
 Authors Authors | Desai N / Brown A | ||||||||||||||||||
| Funding support |  United Kingdom, 5 items United Kingdom, 5 items
| ||||||||||||||||||
 Citation Citation |  Journal: Science / Year: 2017 Journal: Science / Year: 2017Title: The structure of the yeast mitochondrial ribosome. Authors: Nirupa Desai / Alan Brown / Alexey Amunts / V Ramakrishnan /   Abstract: Mitochondria have specialized ribosomes (mitoribosomes) dedicated to the expression of the genetic information encoded by their genomes. Here, using electron cryomicroscopy, we have determined the ...Mitochondria have specialized ribosomes (mitoribosomes) dedicated to the expression of the genetic information encoded by their genomes. Here, using electron cryomicroscopy, we have determined the structure of the 75-component yeast mitoribosome to an overall resolution of 3.3 angstroms. The mitoribosomal small subunit has been built de novo and includes 15S ribosomal RNA (rRNA) and 34 proteins, including 14 without homologs in the evolutionarily related bacterial ribosome. Yeast-specific rRNA and protein elements, including the acquisition of a putatively active enzyme, give the mitoribosome a distinct architecture compared to the mammalian mitoribosome. At an expanded messenger RNA channel exit, there is a binding platform for translational activators that regulate translation in yeast but not mammalian mitochondria. The structure provides insights into the evolution and species-specific specialization of mitochondrial translation. | ||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_3551.map.gz emd_3551.map.gz | 127.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-3551-v30.xml emd-3551-v30.xml emd-3551.xml emd-3551.xml | 108.6 KB 108.6 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_3551.png emd_3551.png | 202.8 KB | ||
| Filedesc metadata |  emd-3551.cif.gz emd-3551.cif.gz | 20.8 KB | ||
| Others |  emd_3551_half_map_1.map.gz emd_3551_half_map_1.map.gz emd_3551_half_map_2.map.gz emd_3551_half_map_2.map.gz | 107.5 MB 107.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-3551 http://ftp.pdbj.org/pub/emdb/structures/EMD-3551 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3551 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3551 | HTTPS FTP |
-Related structure data
| Related structure data |  5mrcMC  3552C  3553C  3554C  3555C  3556C  5mreC  5mrfC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-10107 (Title: The Structure of the Yeast Mitochondrial Ribosome / Data size: 138.6 EMPIAR-10107 (Title: The Structure of the Yeast Mitochondrial Ribosome / Data size: 138.6 Data #1: Yeast mitochondrial ribosome - shiny particles [picked particles - multiframe - processed]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_3551.map.gz / Format: CCP4 / Size: 137.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_3551.map.gz / Format: CCP4 / Size: 137.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.34 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
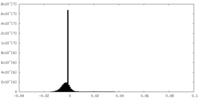
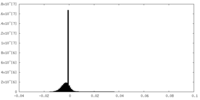
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Half map: #1
| File | emd_3551_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
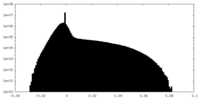
| Density Histograms |
-Half map: #2
| File | emd_3551_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
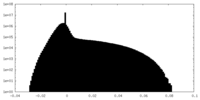
| Density Histograms |
- Sample components
Sample components
+Entire : yeast mitochondrial ribosome - Class A
| Entire | Name: yeast mitochondrial ribosome - Class A |
|---|---|
| Components |
|
+Supramolecule #1: yeast mitochondrial ribosome - Class A
| Supramolecule | Name: yeast mitochondrial ribosome - Class A / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#78 |
|---|---|
| Source (natural) | Organism:  |
+Macromolecule #1: 21S ribosomal RNA
| Macromolecule | Name: 21S ribosomal RNA / type: rna / ID: 1 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 1.057789375 MDa |
| Sequence | String: GUAAAAAGUA GAAUAAUAGA UUUGAAAUAU UUAUUAUAUA GAUUUAAAGA GAUAAUCAUG GAGUAUAAUA AUUAAAUUUA AUAAAUUUA AUAUAACUAU UAAUAGAAUU AGGUUACUAA UAAAUUAAUA ACAAUUAAUU UUAAAACCUA AAGGUAAACC U UUAUAUUA ...String: GUAAAAAGUA GAAUAAUAGA UUUGAAAUAU UUAUUAUAUA GAUUUAAAGA GAUAAUCAUG GAGUAUAAUA AUUAAAUUUA AUAAAUUUA AUAUAACUAU UAAUAGAAUU AGGUUACUAA UAAAUUAAUA ACAAUUAAUU UUAAAACCUA AAGGUAAACC U UUAUAUUA AUAAUGUUAU UUUUUAUUAU UUUUAUAAUA AGAAUAAUUA UUAAUAAUAA UAAACUAAGU GAACUGAAAC AU CUAAGUA ACUUAAGGAU AAGAAAUCAA CAGAGAUAUU AUGAGUAUUG GUGAGAGAAA AUAAUAAAGG UCUAAUAAGU AUU AUGUGA AAAAAAUGUA AGAAAAUAGG AUAACAAAUU CUAAGACUAA AUACUAUUAA UAAGUAUAGU AAGUACCGUA AGGG AAAGU AUGAAAAUGA UUAUUUUAUA AGCAAUCAUG AAUAUAUUAU AUUAUAUUAA UGAUGUACCU UUUGUAUAAU GGGUC AGCA AGUAAUUAAU AUUAGUAAAA CAAUAAGUUA UAAAUAAAUA GAAUAAUAUA UAUAUAUAAA AAAAUAUAUU AAAAUA UUU AAUUAAUAUU AAUUGACCCG AAAGCAAACG AUCUAACUAU GAUAAGAUGG AUAAACGAUC GAACAGGUUG AUGUUGC AA UAUCAUCUGA UUAAUUGUGG UUAGUAGUGA AAGACAAAUC UGGUUUGCAG AUAGCUGGUU UUCUAUGAAA UAUAUGUA A GUAUAGCCUU UAUAAAUAAU AAUUAUUAUA UAAUAUUAUA UUAAUAUUAU AUAAAGAAUG GUACAGCAAU UAAUAUAUA UUAGGGAACU AUUAAAGUUU UAUUAAUAAU AUUAAAUCUC GAAAUAUUUA AUUAUAUAUA AUAAAGAGUC AGAUUAUGUG CGAUAAGGU AAAUAAUCUA AAGGGAAACA GCCCAGAUUA AGAUAUAAAG UUCCUAAUAA AUAAUAAGUG AAAUAAAUAU U AAAAUAUU AUAAUAUAAU CAGUUAAUGG GUUUGACAAU AACCAUUUUU UAAUGAACAU GUAACAAUGC ACUGAUUUAU AA UAAAUAA AAAAAAAUAA UAUUUAAAAU CAAAUAUAUA UAUAUUUGUU AAUAGAUAAU AUACGGAUCU UAAUAAUAAG AAU UAUUUA AUUCCUAAUA UGGAAUAUUA UAUUUUUAUA AUAAAAAUAU AAAUACUGAA UAUCUAAAUA UUAUUAUUAC UUUU UUUUU AAUAAUAAUA AUAUGGUAAU AGAACAUUUA AUGAUAAUAU AUAUUAGUUA UUAAUUAAUA UAUGUAUUAA UUAAA UAGA GAAUGCUGAC AUGAGUAACG AAAAAAAGGU AUAAACCUUU UCACCUAAAA CAUAAGGUUU AACUAUAAAA GUACGG CCC CUAAUUAAAU UAAUAAAAAU AUAAAUAUAU UUAAGAUGGG AUAAUCUAUA UUAAUAAAAA UUUAUCUUAA AAUAUAU AU AUUAUUAAUA AUUAUAUUAA UUAAUUAAUA AUAUAUAUAA UUAUAUUAUA UAUUAUAUAU UUUUUAUAUA AUAUAAAC U AAUAAAGAUC AGGAAAUAAU UAAUGUAUAC CGUAAUGUAG ACCGACUCAG GUAUGUAAGU AGAGAAUAUG AAGGUGAAU UAGAUAAUUA AAGGGAAGGA ACUCGGCAAA GAUAGCUCAU AAGUUAGUCA AUAAAGAGUA AUAAGAACAA AGUUGUACAA CUGUUUACU AAAAACACCG CACUUUGCAG AAACGAUAAG UUUAAGUAUA AGGUGUGAAC UCUGCUCCAU GCUUAAUAUA U AAAUAAAA UUAUUUAACG AUAAUUUAAU UAAAUUUAGG UAAAUAGCAG CCUUAUUAUG AGGGUUAUAA UGUAGCGAAA UU CCUUGGC CUAUAAUUGA GGUCCCGCAU GAAUGACGUA AUGAUACAAC AACUGUCUCC CCUUUAAGCU AAGUGAAAUU GAA AUCGUA GUGAAGAUGC UAUGUACCUU CAGCAAGACG GAAAGACCCU AUGCAGCUUU ACUGUAAUUA GAUAGAUCGA AUUA UUGUU UAUUAUAUUC AGCAUAUUAA GUAAUCCUAU UAUUAGGUAA UCGUUUAGAU AUUAAUGAGA UACUUAUUAU AAUAU AAUG AUAAUUCUAA UCUUAUAAAU AAUUAUUAUU AUUAUUAUUA AUAAUAAUAA UAUGCUUUCA AGCAUAGUGA UAAAAC AUA UUUAUAUGAU AAUCACUUUA CUUAAUAGAU AUAAUUCUUA AGUAAUAUAU AAUAUAUAUU UUAUAUAUAU UAUAUAU AA UAUAAGAGAC AAUCUCUAAU UGGUAGUUUU GAUGGGGCGU CAUUAUCAGC AAAAGUAUCU GAAUAAGUCC AUAAAUAA A UAUAUAAAAU UAUUGAAUAA AAAAAAAAUA AUAUAUAUUA UAUAUAUUAA UUAUAAAUUG AAAUAUGUUU AUAUAAAUU UAUAUUUAUU GAAUAUAUUU UAGUAAUAGA UAAAAAUAUG UACAGUAAAA UUGUAAGGAA AACAAUAAUA ACUUUCUCCU CUCUCGGUG GGGGUUCACA CCUAUUUUUA AUAGGUGUGA ACCCCUCUUC GGGGUUCCGG UUCCCUUUCG GGUCCCGGAA C UUAAAUAA AAAUGGAAAG AAUUAAAUUA AUAUAAUGGU AUAACUGUGC GAUAAUUGUA ACACAAACGA GUGAAACAAG UA CGUAAGU AUGGCAUAAU GAACAAAUAA CACUGAUUGU AAAGGUUAUU GAUAACGAAU AAAAGUUACG CUAGGGAUAA CAG GGUAAU AUAGCGAAAG AGUAGAUAUU GUAAGCUAUG UUUGCCACCU CGAUGUCGAC UCAACAUUUC CUCUUGGUUG UAAA AGCUA AGAAGGGUUU GACUGUUCGU CAAUUAAAAU GUUACGUGAG UUGGGUUAAA UACGAUGUGA AUCAGUAUGG UUCCU AUCU GCUGAAGGAA AUAUUAUCAA AUUAAAUCUC AUUAUUAGUA CGCAAGGACC AUAAUGAAUC AACCCAUGGU GUAUCU AUU GAUAAUAAUA UAAUAUAUUU AAUAAAAAUA AUACUUUAUU AAUAUAUUAU CUAUAUUAGU UUAUAUUUUA AUUAUAU AU UAUCAUAGUA GAUAAGCUAA GUUGAUAAUA AAUAAAUAUU GAAUACAUAU UAAAUAUGAA GUUGUUUUAA UAAGAUAA U UAAUCUGAUA AUUUUAUACU AAAAUUAAUA AUUAUAGGUU UUAUAUAUUA UUUAUAAAUA AAUAUAUUAU AAUAAUAAU AAUUAUUAUU AUUAAUAAAA AAUAUUAAUU AUAAUAUUAA UAAAAUACUA AUUUAUCAGU UAUCUAUAUA AUAUCUAAUC UAUUAUUCU AUAUACU |
+Macromolecule #75: 15S ribosomal RNA
| Macromolecule | Name: 15S ribosomal RNA / type: rna / ID: 75 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 528.784125 KDa |
| Sequence | String: GUAAAAAAUU UAUAAGAAUA UGAUGUUGGU UCAGAUUAAG CGCUAAAUAA GGACAUGACA CAUGCGAAUC AUACGUUUAU UAUUGAUAA GAUAAUAAAU AUGUGGUGUA AACGUGAGUA AUUUUAUUAG GAAUUAAUGA ACUAUAGAAU AAGCUAAAUA C UUAAUAUA ...String: GUAAAAAAUU UAUAAGAAUA UGAUGUUGGU UCAGAUUAAG CGCUAAAUAA GGACAUGACA CAUGCGAAUC AUACGUUUAU UAUUGAUAA GAUAAUAAAU AUGUGGUGUA AACGUGAGUA AUUUUAUUAG GAAUUAAUGA ACUAUAGAAU AAGCUAAAUA C UUAAUAUA UUAUUAUAUA AAAAUAAUUU AUAUAAUAAA AAGGAUAUAU AUAUAAUAUA UAUUUAUCUA UAGUCAAGCC AA UAAUGGU UUAGGUAGUA GGUUUAUUAA GAGUUAAACC UAGCCAACGA UCCAUAAUCG AUAAUGAAAG UUAGAACGAU CAC GUUGAC UCUGAAAUAU AGUCAAUAUC UAUAAGAUAC AGCAGUGAGG AAUAUUGGAC AAUGAUCGAA AGAUUGAUCC AGUU ACUUA UUAGGAUGAU AUAUAAAAAU AUUUUAUUUU AUUUAUAAAU AUUAAAUAUU UAUAAUAAUA AUAAUAAUAA UAUAU AUAU AUAAAUUGAU UAAAAAUAAA AUCCAUAAAU AAUUAAAAUA AUGAUAUUAA UUACCAUAUA UAUUUUUAUA UGGAUA UAU AUAUUAAUAA UAAUAUUAAU UUUAUUAUUA UUAAUAAUAU AUUUUAAUAG UCCUGACUAA UAUUUGUGCC AGCAGUC GC GGUAACACAA AGAGGGCGAG CGUUAAUCAU AAUGGUUUAA AGGAUCCGUA GAAUGAAUUA UAUAUUAUAA UUUAGAGU U AAUAAAAUAU AAUUAAAGAA UUAUAAUAGU AAAGAUGAAA UAAUAAUAAU AAUUAUAAGA CUAAUAUAUG UGAAAAUAU UAAUUAAAUA UUAACUGACA UUGAGGGAUU AAAACUAGAG UAGCGAAACG GAUUCGAUAC CCGUGUAGUU CUAGUAGUAA ACUAUGAAU ACAAUUAUUU AUAAUAUAUA UUAUAUAUAA AUAAUAAAUG AAAAUGAAAG UAUUCCACCU GAAGAGUACG U UAGCAAUA AUGAAACUCA AAACAAUAGA CGGUUACAGA CUUAAGCAGU GGAGCAUGUU AUUUAAUUCG AUAAUCCACG AC UAACCUU ACCAUAUUUU GAAUAUUAUA AUAAUUAUUA UAAUUAUUAU AUUACAGGCG UUACAUUGUU GUCUUUAGUU CGU GCUGCA AAGUUUUAGA UUAAGUUCAU AAACGAACAA AACUCCAUAU AUAUAAUUUU AAUUAUAUAU AAUUUUAUAU UAUU UAUUA AUAUAAAGAA AGGAAUUAAG ACAAAUCAUA AUGAUCCUUA UAAUAUGGGU AAUAGACGUG CUAUAAUAAA AUGAU AAUA AAAUUAUAUA AAAUAUAUUU AAUUAUAUUU AAUUAAUAAU AUAAAACAUU UUAAUUUUUA AUAUAUUUUU UUAUUA UAU AUUAAUAUGA AUUAUAAUCU GAAAUUCGAU UAUAUGAAAA AAGAAUUGCU AGUAAUACGU AAAUUAGUAU GUUACGG UG AAUAUUCUAA CUGUUUCGCA CUAAUCACUC AUCACGCGUU GAAACAUAUU AUUAUCUUAU UAUUUAUAUA AUAUUUUU U AAUAAAUAUU AAUAAUUAUU AAUUUAUAUU UAUUUAUAUC AGAAAUAAUA UGAAUUAAUG CGAAGUUGAA AUACAGUUA CCGUAGGGGA ACCUGCGGUG GGCUUAUAAA UAUCUUAAAU AUUCUUACA |
+Macromolecule #76: tRNA
| Macromolecule | Name: tRNA / type: rna / ID: 76 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 24.422523 KDa |
| Sequence | String: GGCUACGUAG CUCAGUUGGU AGAGCACAUC ACUCAUAAUG AUGGGGUCAC AGGUUCGAAU CCCGUCGUAG CCACCA |
+Macromolecule #2: uL2m
| Macromolecule | Name: uL2m / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 43.870023 KDa |
| Sequence | String: MLVLGSLRSA LSCSSTASLI SKRNPCYPYG ILCRTLSQSV KLWQENTSKD DSSLNITPRL LKIIPNDTDI VTLEKQDELI KRRRKLSKE VTQMKRLKPV SPGLRWYRSP IYPYLYKGRP VRALTVVRKK HGGRNNSGKI TVRHQGGGHR NRTRLIDFNR W EGGAQTVQ ...String: MLVLGSLRSA LSCSSTASLI SKRNPCYPYG ILCRTLSQSV KLWQENTSKD DSSLNITPRL LKIIPNDTDI VTLEKQDELI KRRRKLSKE VTQMKRLKPV SPGLRWYRSP IYPYLYKGRP VRALTVVRKK HGGRNNSGKI TVRHQGGGHR NRTRLIDFNR W EGGAQTVQ RIEYDPGRSS HIALLKHNTT GELSYIIACD GLRPGDVVES FRRGIPQTLL NEMGGKVDPA ILSVKTTQRG NC LPISMIP IGTIIHNVGI TPVGPGKFCR SAGTYARVLA KLPEKKKAIV RLQSGEHRYV SLEAVATIGV VSNIDHQNRS LGK AGRSRW LGIRPTVRGV AMNKCDHPHG GGRGKSKSNK LSMSPWGQLA KGYKTRRGKN QNRMKVKDRP RGKDARL UniProtKB: Large ribosomal subunit protein uL2m |
+Macromolecule #3: uL3m
| Macromolecule | Name: uL3m / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 27.528809 KDa |
| Sequence | String: STRPFLVAPS IANSITTEAP AINHSPELAN ARKWLPKRCG LITRKKGMMP YFDKSTGERS AATILEVNNV EVIMHRTSEV NGYFACQVG YGSRHLSKVS RQMLGHFASK VVNPKEHVAE FRVKDEKGLI PPGTLLKPSF FKEGQYVDVR SVSKGKGFTG V MKRYGFKG ...String: STRPFLVAPS IANSITTEAP AINHSPELAN ARKWLPKRCG LITRKKGMMP YFDKSTGERS AATILEVNNV EVIMHRTSEV NGYFACQVG YGSRHLSKVS RQMLGHFASK VVNPKEHVAE FRVKDEKGLI PPGTLLKPSF FKEGQYVDVR SVSKGKGFTG V MKRYGFKG LRASHGTSIM HRHGGSYGQN QDPGRVLPGR KMPGHMGNEH VTIQNVKVLK VDDENNVIWV KGSVAGPKNS FV KIQDAIK KT UniProtKB: Large ribosomal subunit protein uL3m |
+Macromolecule #4: uL4m
| Macromolecule | Name: uL4m / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 28.254342 KDa |
| Sequence | String: LNPLPNAAIP PKYALVTVRS FPSLEPLTFV PVPTSTVAAP LRRDILWRAV VYENDNRRVG ASNPPGRSEN GFSRRKLMPQ KGSGRARVG DANSPTRHNG GRALARTAPN DYTTELPSKV YSMAFNNALS HQYKSGKLFV IGGEKVDLIS PTPELDLNRL D LVNTNTVE ...String: LNPLPNAAIP PKYALVTVRS FPSLEPLTFV PVPTSTVAAP LRRDILWRAV VYENDNRRVG ASNPPGRSEN GFSRRKLMPQ KGSGRARVG DANSPTRHNG GRALARTAPN DYTTELPSKV YSMAFNNALS HQYKSGKLFV IGGEKVDLIS PTPELDLNRL D LVNTNTVE GKEIFEGEVI FRKFLEEFQL KGKRLLFITD KTREGLIKSS DPYKQKVDVI QKELVEVNDI LRAQAVFIEL EA LEYLAMA HQKEI UniProtKB: Large ribosomal subunit protein uL4m |
+Macromolecule #5: uL5m
| Macromolecule | Name: uL5m / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 31.014529 KDa |
| Sequence | String: PKSACSLVKP VHHLVKIDKS KLSPRFPELK YDKSDIRSPG FKPKDTHADR LNDHYLNTLQ SDLLLINYSH NAAVVKGLKQ RAWSGDSPY HLNRPPKNPR GSKAQLPDIH PIKWSNIPGL ESVVINCFVR EARENQLLAI TAALQLQQIT GCKPHPIFSK N DVPTWKLR ...String: PKSACSLVKP VHHLVKIDKS KLSPRFPELK YDKSDIRSPG FKPKDTHADR LNDHYLNTLQ SDLLLINYSH NAAVVKGLKQ RAWSGDSPY HLNRPPKNPR GSKAQLPDIH PIKWSNIPGL ESVVINCFVR EARENQLLAI TAALQLQQIT GCKPHPIFSK N DVPTWKLR KGHQMGAKVE LKGKEMSQFL STLTEIVLPR IREYKGISNQ SGNRFGGISF GLTAEDIKFF PEIDANQDSW PK TFGMHIN INTSAQLDYQ ARTLLSGFQF PFFGEEK UniProtKB: Large ribosomal subunit protein uL5m |
+Macromolecule #6: uL6m
| Macromolecule | Name: uL6m / type: protein_or_peptide / ID: 6 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 21.690119 KDa |
| Sequence | String: SQVGSLPLYI SPEVQVSINA LSMPRIIRKG RTSMNISQNI TVKGPKGELS VEVPDFLHLD KDEKHGKINV TVQNSEDKHQ RSMWGTVRS LINNHIIGVT EGHLAVLRFV GTGYRAQLEN DGKFVNVKVG ASIKQGLDVP EGIVVKTPAP TSLIIEGCNK Q QVLLFAAK ...String: SQVGSLPLYI SPEVQVSINA LSMPRIIRKG RTSMNISQNI TVKGPKGELS VEVPDFLHLD KDEKHGKINV TVQNSEDKHQ RSMWGTVRS LINNHIIGVT EGHLAVLRFV GTGYRAQLEN DGKFVNVKVG ASIKQGLDVP EGIVVKTPAP TSLIIEGCNK Q QVLLFAAK LRKFHPPEPY KGKGIYVNDE TIKLKDKK UniProtKB: Large ribosomal subunit protein uL6m |
+Macromolecule #7: bL9m
| Macromolecule | Name: bL9m / type: protein_or_peptide / ID: 7 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 8.763024 KDa |
| Sequence | String: SALTKRTHRV KVQVLKDFPR FQLYKGQVAN VKPSLMRNYL HNFNGAKYIL SEEHDINTEL LKQYQTLEAK LEED UniProtKB: Large ribosomal subunit protein bL9m |
+Macromolecule #8: uL13m
| Macromolecule | Name: uL13m / type: protein_or_peptide / ID: 8 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 18.125967 KDa |
| Sequence | String: SQKIGHSGLA FARLWHHVDV ARDKRTLGRL ASAIAITLIG RHKPVYHPSQ DCGDYVVVTN CQKIRVTGKK FEQKTYWSHS GRPGQLKLQ TMNKVVADKG FGEILKKAVS GMLPKNKLRK QRLDRLKVFD GSENPYKQNI TAFAHEQSSI PEPLKESIFN Q UniProtKB: Large ribosomal subunit protein uL13m |
+Macromolecule #9: uL14m
| Macromolecule | Name: uL14m / type: protein_or_peptide / ID: 9 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 14.929664 KDa |
| Sequence | String: MIFLKSVIKV IDNSGAQLAE CIKVIRKGSP KSPAMVGDRI VCVIQKAKPL TQNITGTANT NRVKKGDICH AIVVRSKQRN MCRKDGSTV AFGDTACVLI NKNTGEPLGT RIMANDGCVD RTLKDKGYNK ICSLASRVI UniProtKB: Large ribosomal subunit protein uL14m |
+Macromolecule #10: uL15m
| Macromolecule | Name: uL15m / type: protein_or_peptide / ID: 10 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 24.765012 KDa |
| Sequence | String: VSILGQLKPS DGSTKSFKRL GRGPSSGLGK TSGRGQKGQK ARGKVKSWFE GGQTPIYKLF PKIGFTNVGA KPLKELNLKR IQWFHDKNR LHLQPGEVLD MNKMRKLGLV TGPIKYGVKI LASGKFHYNL PIALEASRAS AKAIAAIEKA GGKFTARYYT P LGLRAHLN ...String: VSILGQLKPS DGSTKSFKRL GRGPSSGLGK TSGRGQKGQK ARGKVKSWFE GGQTPIYKLF PKIGFTNVGA KPLKELNLKR IQWFHDKNR LHLQPGEVLD MNKMRKLGLV TGPIKYGVKI LASGKFHYNL PIALEASRAS AKAIAAIEKA GGKFTARYYT P LGLRAHLN PQWFLEKRGR VPLQARPTKR RDIDFYSKEE KRGYLVMEKD KLLQDIKEAQ NK UniProtKB: Large ribosomal subunit protein uL15m |
+Macromolecule #11: uL16m
| Macromolecule | Name: uL16m / type: protein_or_peptide / ID: 11 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 22.347031 KDa |
| Sequence | String: KHEYAPRFKI VQKKQKGRVP VRTGGSIKGS TLQFGKYGLR LKSEGIRISA QQLKEADNAI MRYVRPLNNG HLWRRLCTNV AVCIKGNET RMGKGKGGFD HWMVRVPTGK ILFEINGDDL HEKVAREAFR KAGTKLPGVY EFVSLDSLVR VGLHSFKNPK D DPVKNFYD ...String: KHEYAPRFKI VQKKQKGRVP VRTGGSIKGS TLQFGKYGLR LKSEGIRISA QQLKEADNAI MRYVRPLNNG HLWRRLCTNV AVCIKGNET RMGKGKGGFD HWMVRVPTGK ILFEINGDDL HEKVAREAFR KAGTKLPGVY EFVSLDSLVR VGLHSFKNPK D DPVKNFYD ENAKKPSKKY LNILKSQEPQ YKLFRGR UniProtKB: Large ribosomal subunit protein uL16m |
+Macromolecule #12: bL17m
| Macromolecule | Name: bL17m / type: protein_or_peptide / ID: 12 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 26.858182 KDa |
| Sequence | String: TVGIARKLSR DKAHRDALLK NLACQLFQHE SIVSTHAKCK EASRVAERII TWTKRAITTS NSVAQAELKS QIQSQLFLAG DNRKLMKRL FSEIAPRYLE RPGGYTRVLR LEPRANDSAP QSVLELVDSP VMSESHTVNR GNLKMWLLVK SVINDDANQL P HNPLTLQN ...String: TVGIARKLSR DKAHRDALLK NLACQLFQHE SIVSTHAKCK EASRVAERII TWTKRAITTS NSVAQAELKS QIQSQLFLAG DNRKLMKRL FSEIAPRYLE RPGGYTRVLR LEPRANDSAP QSVLELVDSP VMSESHTVNR GNLKMWLLVK SVINDDANQL P HNPLTLQN LHKVAKFKAE AQLHGEIMLI KQVLLKEMSL PYDEALENER TQALLKEVYS SSLPKKTKKP SSYVMVPRP UniProtKB: Large ribosomal subunit protein bL17m |
+Macromolecule #13: bL19m
| Macromolecule | Name: bL19m / type: protein_or_peptide / ID: 13 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 17.179088 KDa |
| Sequence | String: YMVPATKRKT IPVYPPVQRI ASSQIMKQVA LSEIESLDPG AVKRKLISKK NKDRLKAGDV VRIVYDSSKC SYDTFVGYIL SIDRKQLVQ DASLLLRNQI AKTAVEIRVP LFSPLIERID LLTPHVSSRQ RNKHYYIRGT RLDVGDLEAG LR UniProtKB: Large ribosomal subunit protein bL19m |
+Macromolecule #14: bL21m
| Macromolecule | Name: bL21m / type: protein_or_peptide / ID: 14 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 13.507729 KDa |
| Sequence | String: TDTTPLKLSN ELYAIFKIHN RPYLVTEGDR VILPFKLKQA EVGDILNMTD VTTLGSRNYK LVGHPINTSL YTLKATVVGK TKRAFQTRE VTKRRNRRVR HAKSKGDLTI LRISELSMN UniProtKB: Large ribosomal subunit protein bL21m |
+Macromolecule #15: uL22m
| Macromolecule | Name: uL22m / type: protein_or_peptide / ID: 15 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 25.964479 KDa |
| Sequence | String: SITIENDKLL QQHIISLQQP EQLASQSLLS PLKREIYEAN CKINGGFYKK DTIVKLPNSS ERYKLKLTKR EIEVLEPSVY AQSYRIKSS MKKATLLLRL LGGLDVMKAI SQCHFSNKKI AREVAELLQK GVKDGQKLGL KPEDLYISQI WTGSDGFWRK R VEFKARTR ...String: SITIENDKLL QQHIISLQQP EQLASQSLLS PLKREIYEAN CKINGGFYKK DTIVKLPNSS ERYKLKLTKR EIEVLEPSVY AQSYRIKSS MKKATLLLRL LGGLDVMKAI SQCHFSNKKI AREVAELLQK GVKDGQKLGL KPEDLYISQI WTGSDGFWRK R VEFKARTR IGIISHPYIH VRCILRTKSV TKRRLAYEAH LKEQKRAPWV QLGDKPIRGV TGGVYKW UniProtKB: Large ribosomal subunit protein uL22m |
+Macromolecule #16: uL23m
| Macromolecule | Name: uL23m / type: protein_or_peptide / ID: 16 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 24.529098 KDa |
| Sequence | String: DINVSEKIYK WTKAGIEQGK EHFKVGGNKV YFPKARIILL RPNAKHTPYQ AKFIVPKSFN KLDLRDYLYH IYGLRAMNIT TQLLHGKFN RMNLQTTRFR EPQIKKMTIE MEEPFIWPEE PRPDENSFWD STTPDNMEKY REERLNCLGS DANKPGTAFD G VVGPYERV ...String: DINVSEKIYK WTKAGIEQGK EHFKVGGNKV YFPKARIILL RPNAKHTPYQ AKFIVPKSFN KLDLRDYLYH IYGLRAMNIT TQLLHGKFN RMNLQTTRFR EPQIKKMTIE MEEPFIWPEE PRPDENSFWD STTPDNMEKY REERLNCLGS DANKPGTAFD G VVGPYERV AQPFIPRFLK REIDNKRERH AAELQRADKL IALNRYIED UniProtKB: Large ribosomal subunit protein uL23m |
+Macromolecule #17: uL24m
| Macromolecule | Name: uL24m / type: protein_or_peptide / ID: 17 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 33.67684 KDa |
| Sequence | String: SGSYQHLSNV GSRVMKRLGN RPKNFLPHSE KFIKKSTPEF MKSDLKEVDE KTSFKSEKEW KFIPGDRVVV MSGASKGNIA VIKSFDKRT NSFILDENGP TKTVPVPKQF WLEGQTSHMI TIPVSILGKD LRLVADIDDE KTPGKTRTVA VRDVSFNGSY Y DADYKKVM ...String: SGSYQHLSNV GSRVMKRLGN RPKNFLPHSE KFIKKSTPEF MKSDLKEVDE KTSFKSEKEW KFIPGDRVVV MSGASKGNIA VIKSFDKRT NSFILDENGP TKTVPVPKQF WLEGQTSHMI TIPVSILGKD LRLVADIDDE KTPGKTRTVA VRDVSFNGSY Y DADYKKVM PYRCVKGQPD LIIPWPKPDP IDVQTNLATD PVIAREQTFW VDSVVRNPIP KKAIPSIRNP HSKYKRGTLT AK DIAKLVA PEMPLTEVRK SHLAEKKELA EREVPKLTEE DMEAIGARVF EFLEKQKRE UniProtKB: Large ribosomal subunit protein uL24m |
+Macromolecule #18: bL27m
| Macromolecule | Name: bL27m / type: protein_or_peptide / ID: 18 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 39.516336 KDa |
| Sequence | String: SRTSMKDSAG RRLGPKKYEG QDVSTGEIIM RQRGTKFYPG ENVGIGKDHS IFALEPGVVR YYLDPFHPKR KFIGVALRRD LKLPSPHFE PTVRRFGRFE LTNKRAAYKE ENSISRKDYL AKPNILKQLE VRESKRKELQ DKLSKVLRDE LKLDIKDIEL A TSYLIRVR ...String: SRTSMKDSAG RRLGPKKYEG QDVSTGEIIM RQRGTKFYPG ENVGIGKDHS IFALEPGVVR YYLDPFHPKR KFIGVALRRD LKLPSPHFE PTVRRFGRFE LTNKRAAYKE ENSISRKDYL AKPNILKQLE VRESKRKELQ DKLSKVLRDE LKLDIKDIEL A TSYLIRVR ASLKNGYPIE DARFNSRYYL KEEERLKARR ESWTNEKLSE SLSKIDECSD LLNSSTSFNN KLELHQYISE QE KQALKAK LLEDLEKSQH LETKKDKNYI KALFKDACNF LTLSEEVHLR RKYLKSVFPE TDSTVETKSG KKSIVSRRFD YTK NKVEVI ARSRRAFLSK L UniProtKB: Large ribosomal subunit protein bL27m |
+Macromolecule #19: bL28m
| Macromolecule | Name: bL28m / type: protein_or_peptide / ID: 19 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 25.193494 KDa |
| Sequence | String: RQWRLIETRK IAKQPNYQVG DAKPLHMPKE RKKFPDYKYG ESNIFKQSNK GLYGGSFVQF GNNISESKAK TRKKWLPNVV KKGLWSETL NRKISIKMTA KVLKTISKEG GIDNYLTKEK SARIKELGPT GWKLRYRVLK RKDEIENPPH KDAPIIEMAG G KKAKIYYD ...String: RQWRLIETRK IAKQPNYQVG DAKPLHMPKE RKKFPDYKYG ESNIFKQSNK GLYGGSFVQF GNNISESKAK TRKKWLPNVV KKGLWSETL NRKISIKMTA KVLKTISKEG GIDNYLTKEK SARIKELGPT GWKLRYRVLK RKDEIENPPH KDAPIIEMAG G KKAKIYYD EIVNGSPRKI SVGRRRLMSF LYPLEKLEYR SVGKDLNYKK FVELFADV UniProtKB: Large ribosomal subunit protein bL28m |
+Macromolecule #20: uL29m
| Macromolecule | Name: uL29m / type: protein_or_peptide / ID: 20 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 26.410955 KDa |
| Sequence | String: ARTKFTKPKP KQPVLPKDKI RPPTQLTHHS NNLRITEPIP PTTSNLRCPD DHPLWQFFSN KKFIRSADDL PPSSHIRPWS IPELRHKSF NDLHSLWYNC LREQNVLARE NHLLKNIVGS THDEFSELSN SIRTTMWQIR HVLNERELAY SASREFLQDE S ERKKFLDT ...String: ARTKFTKPKP KQPVLPKDKI RPPTQLTHHS NNLRITEPIP PTTSNLRCPD DHPLWQFFSN KKFIRSADDL PPSSHIRPWS IPELRHKSF NDLHSLWYNC LREQNVLARE NHLLKNIVGS THDEFSELSN SIRTTMWQIR HVLNERELAY SASREFLQDE S ERKKFLDT LANDYFLNKD IPDDEVASML TRFQLAIFGI SETIQDNTVD INFIDGIKFL ANLKLQR UniProtKB: Large ribosomal subunit protein uL29m |
+Macromolecule #21: uL30m
| Macromolecule | Name: uL30m / type: protein_or_peptide / ID: 21 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 9.088729 KDa |
| Sequence | String: VFYKVTLSRS LIGVPHTTKS IVKSLGLGKR GSIVYKKVNP AIAGSLAKVK ELVKVEVTEH ELTPSQQREL RKSNPGFIVE KR UniProtKB: Large ribosomal subunit protein uL30m |
+Macromolecule #22: bL31m
| Macromolecule | Name: bL31m / type: protein_or_peptide / ID: 22 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 20.125756 KDa |
| Sequence | String: MLKSIFAKRF ASTGSYPGST RITLPRRPAK KIQLGKSRPA IYHQFNVKME LSDGSVVIRR SQYPKGEIRL IQDQRNNPLW NPSRDDLVV VDANSGGSLD RFNKRYSSLF SVDSTTPNSS SETVELSEEN KKKTQIKKEE KEDVSEKAFG MDDYLSLLDD S EQQIKSGK LASKKRDKK UniProtKB: Large ribosomal subunit protein bL31m |
+Macromolecule #23: bL32m
| Macromolecule | Name: bL32m / type: protein_or_peptide / ID: 23 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 13.409719 KDa |
| Sequence | String: AVPKKKVSHQ KKRQKLYGPG KKQLKMIHHL NKCPSCGHYK RANTLCMYCV GQISHIWKTH TAKEEIKPRQ EEELSELDQR VLYPGRRDT KYTKDLKDKD NYLERRVRTL KKD UniProtKB: Large ribosomal subunit protein bL32m |
+Macromolecule #24: bL33m
| Macromolecule | Name: bL33m / type: protein_or_peptide / ID: 24 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 7.275654 KDa |
| Sequence | String: SKNSVIKLLS TAASGYSRYI SIKKGAPLVT QVRYDPVVKR HVLFKEAKKR KVAERKPLDF LRTA UniProtKB: Large ribosomal subunit protein bL33m |
+Macromolecule #25: bL34m
| Macromolecule | Name: bL34m / type: protein_or_peptide / ID: 25 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 5.465526 KDa |
| Sequence | String: KSRGNTYQPS TLKRKRTFGF LARAKSKQGS KILKRRKLKG RWFLSH UniProtKB: Large ribosomal subunit protein bL34m |
+Macromolecule #26: bL35m
| Macromolecule | Name: bL35m / type: protein_or_peptide / ID: 26 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 7.206557 KDa |
| Sequence | String: LMKTHKGTAK RWRRTGNTFK RGIAGRKHGN IGWSHRSLKA LTGRKIAHPA YSKHLKRLLP YH UniProtKB: Large ribosomal subunit protein bL35m |
+Macromolecule #27: bL36m
| Macromolecule | Name: bL36m / type: protein_or_peptide / ID: 27 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 4.653594 KDa |
| Sequence | String: FKVRTSVKKF CSDCYLVRRK GRVYIYCKSN KKHKQRQG UniProtKB: Large ribosomal subunit protein bL36m |
+Macromolecule #28: mL38
| Macromolecule | Name: mL38 / type: protein_or_peptide / ID: 28 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 40.703473 KDa |
| Sequence | String: HIWSDFTTRP SSLSIQSSKV KNYLFQKKAS LDPPSISRRS NRIKYSPPEH IDEIFRMSYD FLEQRSSKFY ELANKTKNPL KKDALLIKA EINNPEVQYN FQFNNKLNNV KDIIDYDVPV YRHLGKQHWE SYGQMLLMQR LETLAAIPDT LPTLVPRAEV N IKFPFSTG ...String: HIWSDFTTRP SSLSIQSSKV KNYLFQKKAS LDPPSISRRS NRIKYSPPEH IDEIFRMSYD FLEQRSSKFY ELANKTKNPL KKDALLIKA EINNPEVQYN FQFNNKLNNV KDIIDYDVPV YRHLGKQHWE SYGQMLLMQR LETLAAIPDT LPTLVPRAEV N IKFPFSTG VNKWIEPGEF LSSNVTSMRP IFKIQEYELV NVEKQLYTVL IVNPDVPDLS NDSFKTALCY GLVNINLTYN DN LIDPRKF HSSNIIADYL PPVPEKNAGK QRFVVWVFRQ PLIEDKQGPN MLEIDRKELS RDDFDIRQFT KKYNLTAIGA HIW RSEWDA KVAAVREKYG LPPGRVFSRV RR UniProtKB: Large ribosomal subunit protein mL38 |
+Macromolecule #29: mL40
| Macromolecule | Name: mL40 / type: protein_or_peptide / ID: 29 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 13.464595 KDa |
| Sequence | String: SLSPLAQRVV TQLSVMSASR KQPKLLKLAR EDLIKHQTIE KCWSIYQQQQ RERRNLQLEL QYKSIERSMN LLQELSPRLF EAANASEKG KRFPMEMKVP TDFPPNTLWH YNFR UniProtKB: Large ribosomal subunit protein mL40 |
+Macromolecule #30: mL41
| Macromolecule | Name: mL41 / type: protein_or_peptide / ID: 30 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 14.841158 KDa |
| Sequence | String: LTRPWKKYRD GELFYGLSKV GNKRVPLTTK QGNKTMYKGT RASGIGRHTK FGGYVINWKK VRTYVTPDMV NFELKPYVNA NVPPLKHEF KGFSGGPLDP RLQLLKIKEY IVNGRVQSEG ATDTSCYKER G UniProtKB: Large ribosomal subunit protein mL41 |
+Macromolecule #31: mL43
| Macromolecule | Name: mL43 / type: protein_or_peptide / ID: 31 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 15.909393 KDa |
| Sequence | String: VVKAIARNSI GRNGVGAFVF PCRKITLQFC NWGGSSEGMR KFLTSKRLDK WGQEFPWIQF EVMRKSGHPL LRAEYTNGRE KVICVRNLN IDNVENKLKL LKDSDGDILR RRTKNDNVES LNSSVRGIWS PLHAAKRHR UniProtKB: Large ribosomal subunit protein mL43 |
+Macromolecule #32: mL44
| Macromolecule | Name: mL44 / type: protein_or_peptide / ID: 32 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 36.326758 KDa |
| Sequence | String: ESELAKYKEY YQGLKSTVNE IPESVASKSP SLRTLHKRLQ LPNELTYSTL SRCLTCPSAK LPDKINNPTK GAAFVNTVPT NKYLDNHGL NIMGKNLLSY HVTKSIIQKY PRLPTVVLNA AVNAYISEAV LAHIAKYWGI EVETTSVLSR YLKMEPFEFT L GRLKFFNN ...String: ESELAKYKEY YQGLKSTVNE IPESVASKSP SLRTLHKRLQ LPNELTYSTL SRCLTCPSAK LPDKINNPTK GAAFVNTVPT NKYLDNHGL NIMGKNLLSY HVTKSIIQKY PRLPTVVLNA AVNAYISEAV LAHIAKYWGI EVETTSVLSR YLKMEPFEFT L GRLKFFNN SLNSKDGIEL ITGKNFSETS ALAMSVRSII AAIWAVTEQK DSQAVYRFID DHIMSRKLDI TKMFQFEQPT RE LAMLCRR EGLEKPVSKL VAESGRLSKS PVFIVHVFSG EETLGEGYGS SLKEAKARAA TDALMKWYCY EPLAQQEPVI DPG TVVV UniProtKB: Large ribosomal subunit protein mL44 |
+Macromolecule #33: mL46
| Macromolecule | Name: mL46 / type: protein_or_peptide / ID: 33 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 32.26174 KDa |
| Sequence | String: MKVNLMLKRG LATATATASS APPKIKVGVL LSRIPIIKSE LNELEKKYYE YQSELEKRLM WTFPAYFYFK KGTVAEHKFL SLQKGPISK KNGIWFPRGI PDIKHGRERS TKQEVKLSDD STVAFSNNQK EQSKDDVNRP VIPNDRITEA DRSNDMKSLE R QLSRTLYL ...String: MKVNLMLKRG LATATATASS APPKIKVGVL LSRIPIIKSE LNELEKKYYE YQSELEKRLM WTFPAYFYFK KGTVAEHKFL SLQKGPISK KNGIWFPRGI PDIKHGRERS TKQEVKLSDD STVAFSNNQK EQSKDDVNRP VIPNDRITEA DRSNDMKSLE R QLSRTLYL LVKDKSGTWK FPNFDLSDES KPLHVHAENE LKLLSGDQIY TWSVSATPIG VLQDERNRTA EFIVKSHILA GK FDLVASK NDAFEDFAWL TKGEISEYVP KDYFNKTEFL LADN UniProtKB: Large ribosomal subunit protein mL46 |
+Macromolecule #34: mL49
| Macromolecule | Name: mL49 / type: protein_or_peptide / ID: 34 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 12.182286 KDa |
| Sequence | String: APIFPKLEDV KMHELIGNNN FGKKTYYVER SRTGNLPVYS AYKNGGNKII TEIRKIEGDV IQLRNDLQEQ LPFIPKKSWS VVMQSKKII IKGNAVEAVK RVLTKKF UniProtKB: Large ribosomal subunit protein mL49 |
+Macromolecule #35: mL50
| Macromolecule | Name: mL50 / type: protein_or_peptide / ID: 35 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 30.317365 KDa |
| Sequence | String: MSSLLKLHCI RPLPQRSVWL SGYKQKARCI HSSAANGDFM SWFKRKKQEE HQEPVKDTKQ LIKDIEEGTN EASSQSSSNN KNRLELIPE NFIGEGSRRC KRQKELKLAV SSAPFNQWLS RDKITSDNQL DDMILQATEK TLGKVDQDVQ FSDLVAKFQF T KFLQSKSG ...String: MSSLLKLHCI RPLPQRSVWL SGYKQKARCI HSSAANGDFM SWFKRKKQEE HQEPVKDTKQ LIKDIEEGTN EASSQSSSNN KNRLELIPE NFIGEGSRRC KRQKELKLAV SSAPFNQWLS RDKITSDNQL DDMILQATEK TLGKVDQDVQ FSDLVAKFQF T KFLQSKSG YLIPDYELTT LSTPLQFKRY IKEKILPSAN DPKLAYKEAE PNAIHPFSDN YASPNIYVVN DVTSKEQKSK YD TIMKEIQ KLEDDATRKA LETARSA UniProtKB: Large ribosomal subunit protein mL50 |
+Macromolecule #36: mL57
| Macromolecule | Name: mL57 / type: protein_or_peptide / ID: 36 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 23.816342 KDa |
| Sequence | String: VIYLHKGPRI NGLRRDPESY LRNPSGVLFT EVNAKECQDK VRSILQLPKY GINLSNELIL QCLTHKSFAH GSKPYNEKLN LLGAQFLKL QTCIHSLKNG SPAESCENGQ LSLQFSNLGT KFAKELTSKN TACTFVKLHN LDPFIFWKMR DPIKDGHING E TTIFASVL ...String: VIYLHKGPRI NGLRRDPESY LRNPSGVLFT EVNAKECQDK VRSILQLPKY GINLSNELIL QCLTHKSFAH GSKPYNEKLN LLGAQFLKL QTCIHSLKNG SPAESCENGQ LSLQFSNLGT KFAKELTSKN TACTFVKLHN LDPFIFWKMR DPIKDGHING E TTIFASVL NAFIGAILST NGSEKAAKFI QGSLLDKEDL HSLVNIANEN VASAKAK UniProtKB: Large ribosomal subunit protein mL57 |
+Macromolecule #37: mL58
| Macromolecule | Name: mL58 / type: protein_or_peptide / ID: 37 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 20.507609 KDa |
| Sequence | String: KQFGFPKTQV TTIYNKTKSA SNYKGYLKHR DAPGMYYQPS ESIATGSVNS ETIPRSFMAA SDPRRGLDMP VQSTKAKQCP NVLVGKSTV NGKTYHLGPQ EIDEIRKLRL DNPQKYTRKF LAAKYGISPL FVSMVSKPSE QHVQIMESRL QEIQSRWKEK R RIAREDRK RRKLLWYQA UniProtKB: Large ribosomal subunit protein mL58 |
+Macromolecule #38: mL59
| Macromolecule | Name: mL59 / type: protein_or_peptide / ID: 38 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 18.408705 KDa |
| Sequence | String: YKQYFDSLPL KLKSFFQRYP PSIKYSPVST STKAINANPF LPNKHPVTQR FHDPKYSLRR MSDVYKLALR YGVEEFLPPI ENTKKLFFE EKYNKKTLMK GVLLPKGHKH ELKLNEKLKK REEALKKVDE LIASKKGSKY AKRVEKMKKN QSIGWF UniProtKB: Large ribosomal subunit protein mL59 |
+Macromolecule #39: mL60
| Macromolecule | Name: mL60 / type: protein_or_peptide / ID: 39 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 14.277905 KDa |
| Sequence | String: GGLLWKIPWR MSTHQKTRQR ERLRNVDQVI KQLTLGLHVQ RCQDKGLTYQ EAMESKKKYK PRSKSLRLLN KPSVFPKENQ MSSKDKYWT FDKKAVGYRK GIHKVPKWTK ISIRKAPKFF UniProtKB: Large ribosomal subunit protein mL60 |
+Macromolecule #40: mL67
| Macromolecule | Name: mL67 / type: protein_or_peptide / ID: 40 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 25.712518 KDa |
| Sequence | String: MKVNHSISRF RPASWFEKTK IIPPQVYIFR NLEYGQVLYS QFPNFSQTQV DKLFVRPNWS NRKPSLRRDI WKCMCVVNLQ NYKQSVHLY QNLCRLRYLR DVAQRKESDK LRKKDSNGHV WYSGQYRPTY CQEAVADLRE SLLKVFENAT PAEKQTVPAK K PSIYWEDP ...String: MKVNHSISRF RPASWFEKTK IIPPQVYIFR NLEYGQVLYS QFPNFSQTQV DKLFVRPNWS NRKPSLRRDI WKCMCVVNLQ NYKQSVHLY QNLCRLRYLR DVAQRKESDK LRKKDSNGHV WYSGQYRPTY CQEAVADLRE SLLKVFENAT PAEKQTVPAK K PSIYWEDP WRMGDKDKHW NYDVFNALGL EHKLIQRVGN IAREESVILK ELAKLES UniProtKB: Large ribosomal subunit protein mL67 |
+Macromolecule #41: bS1m
| Macromolecule | Name: bS1m / type: protein_or_peptide / ID: 41 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 39.507457 KDa |
| Sequence | String: MTLAELLGRS RIAQVANNHK PLTYTGKKFH PTHQIIETKP STLYRQEWGL KSAIPSKIKS RYLVYNDLDT LERITTFEPR GGTQWNRLR FQEMGVPIVS NIGRQNPFFK YISRPEDESH AKLSLFKEMK GDTDISPAAM KKRLKKITAL IRSFQDEFKE W LVENHPDE ...String: MTLAELLGRS RIAQVANNHK PLTYTGKKFH PTHQIIETKP STLYRQEWGL KSAIPSKIKS RYLVYNDLDT LERITTFEPR GGTQWNRLR FQEMGVPIVS NIGRQNPFFK YISRPEDESH AKLSLFKEMK GDTDISPAAM KKRLKKITAL IRSFQDEFKE W LVENHPDE LKLNSNKLED YVVKFLNKKL ETKTNKKFNT EIIGTGGLSY SLPGKLKNSP NGVIQRTVVP GRILNVVKEN ND NKWLAAI GGFVADVVFF QSPPSSFNSM GDFIRMKTFL FEILEASMEK NGSVSMHARL LEPQNDKTRE FFNKRPIYKP LTS RRARRP SVGNIQEANN LLNIIKGN UniProtKB: Small ribosomal subunit protein bS1m |
+Macromolecule #42: uS2m
| Macromolecule | Name: uS2m / type: protein_or_peptide / ID: 42 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 29.585344 KDa |
| Sequence | String: PYPNLIPSAN DKPYSSQELF LRQLNHSMRT AKLGATISKV YYPHKDIFYP PLPENITVES LMSAGVHLGQ STSLWRSSTQ SYIYGEYKG IHIIDLNQTL SYLKRAAKVV EGVSESGGII LFLGTRQGQK RGLEEAAKKT HGYYVSTRWI PGTLTNSTEI S GIWEKQEI ...String: PYPNLIPSAN DKPYSSQELF LRQLNHSMRT AKLGATISKV YYPHKDIFYP PLPENITVES LMSAGVHLGQ STSLWRSSTQ SYIYGEYKG IHIIDLNQTL SYLKRAAKVV EGVSESGGII LFLGTRQGQK RGLEEAAKKT HGYYVSTRWI PGTLTNSTEI S GIWEKQEI DSNDNPTERA LSPNETSKQV KPDLLVVLNP TENRNALLEA IKSRVPTIAI IDTDSEPSLV TYPIPGNDDS LR SVNFLLG VLARAGQRGL QNRLARNNE UniProtKB: Small ribosomal subunit protein uS2m |
+Macromolecule #43: uS3m
| Macromolecule | Name: uS3m / type: protein_or_peptide / ID: 43 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 47.170996 KDa |
| Sequence | String: MKLKLLNMIL SMMNKTNNNN NIIINNTLDS LMNKKLLLKN MLLDMNNKKM NNMKRMLNNN NMNPAGANPV VHRIGPAGNI NNKLQHLNN MNNWNTQIYN YNKNMEIMNT MNDKLINKLL YKMMTLKLNN MNINKIIMSK TINQHSLNKL NIKFYYYNND I NNNNNNNN ...String: MKLKLLNMIL SMMNKTNNNN NIIINNTLDS LMNKKLLLKN MLLDMNNKKM NNMKRMLNNN NMNPAGANPV VHRIGPAGNI NNKLQHLNN MNNWNTQIYN YNKNMEIMNT MNDKLINKLL YKMMTLKLNN MNINKIIMSK TINQHSLNKL NIKFYYYNND I NNNNNNNN NNYYMNMMNK LMNIMNNNMN NNLCNILSYY YKKKVTIEPI KLSYIYLNSD IFSKYISLND MDKYNNGILT NY QRMLNNI MPKLNDHNIS MNYINNINNI NNNKYNNMIN LLNNNNNINN NNNYNNNNNN YIGNINNIYN NMTIDNIPMD ILM YKYLVG WSIKFKGRLS NNNGRTSTTN LLNGTFNNKK YLWSNINNNY KLNYIPSNHN LYNNSNINKN GKYNIKVKLN FI UniProtKB: Small ribosomal subunit protein uS3m |
+Macromolecule #44: uS4m
| Macromolecule | Name: uS4m / type: protein_or_peptide / ID: 44 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 56.450949 KDa |
| Sequence | String: MPRKANLLKS LARGRVRTSF NKYNLFNLYK KGGVDLKSKS LYQQKWTAKQ ETRAYHGEHL TEKRWQTVFK PKLDSVAQLD ASLRGGEIK ETPFLLQTFA VLEKRLDFAL FRAMFASSVR QARQFILHGN VRVNGVKIKH PSYTLKPGDM FSVKPDKVLE A LGAKKPSF ...String: MPRKANLLKS LARGRVRTSF NKYNLFNLYK KGGVDLKSKS LYQQKWTAKQ ETRAYHGEHL TEKRWQTVFK PKLDSVAQLD ASLRGGEIK ETPFLLQTFA VLEKRLDFAL FRAMFASSVR QARQFILHGN VRVNGVKIKH PSYTLKPGDM FSVKPDKVLE A LGAKKPSF QEALKIDKTQ IVLWNKYVKE AKTEPKEVWE KKLENFEKMS DSNPKKLQFQ EFLRQYNKNL ESQQYNALKG CT QEGILRK LLNVEKEIGK SNNEPLSIDE LKQGLPEIQD SQLLESLNNA YQEFFKSGEI RREIISKCQP DELISLATEM MNP NETTKK ELSDGAKSAL RSGKRIIAES VKLWTKNITD HFKTRMSDIS DGSLTFDPKW AKNLKYHDPI KLSELEGDEP KARK LINLP WQKNYVYGRQ DPKKPFFTPW KPRPFLSPFA ILPHHLEISF KTCHAVYLRD PVARPGQSEV ISPFDVPVHE RAYMY YLRN GK UniProtKB: Small ribosomal subunit protein uS4m |
+Macromolecule #45: uS5m
| Macromolecule | Name: uS5m / type: protein_or_peptide / ID: 45 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 33.24009 KDa |
| Sequence | String: QHYDESLLSR YYPESLLKSI KLAQQTIPED TKFRVSRNVE FAPPYLDDFT KIHPFWDYKP GMPHLHAQEE NNNFSIFRWD QVQQPLPGE GNILPPGVSL PNDGGRKSKS ADVAAGLHKQ TGVDPDYITR KLTMKPLVMK RVSNQTGKGK IASFYALVVV G DKNGMVGL ...String: QHYDESLLSR YYPESLLKSI KLAQQTIPED TKFRVSRNVE FAPPYLDDFT KIHPFWDYKP GMPHLHAQEE NNNFSIFRWD QVQQPLPGE GNILPPGVSL PNDGGRKSKS ADVAAGLHKQ TGVDPDYITR KLTMKPLVMK RVSNQTGKGK IASFYALVVV G DKNGMVGL GEGKSREEMS KAIFKAHWDA VRNLKEIPRY ENRTIYGDID FRYHGVKLHL RSAKPGFGLR VNHVIFEICE CA GIKDLSG KVYKSRNDMN IAKGTIEAFT KAQKTLDEVA LGRGKKLVDV RKVYYS UniProtKB: Small ribosomal subunit protein uS5m |
+Macromolecule #46: bS6m
| Macromolecule | Name: bS6m / type: protein_or_peptide / ID: 46 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 14.310959 KDa |
| Sequence | String: MLYELIGLVR ITNSNAPKLE AKELSSTIGK LIIQNRGVVR DIVPMGIRYL PKIMKKDQEK HFRAYHFLML FDSSAAVQSE ILRTLKKDP RVIRSSIVKV DLDKQLDRAS SLHRSLGKKS ILELVN UniProtKB: Small ribosomal subunit protein bS6m |
+Macromolecule #47: uS7m
| Macromolecule | Name: uS7m / type: protein_or_peptide / ID: 47 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 18.271279 KDa |
| Sequence | String: KIKPTAEQLA QWEALKSVPI PPRKNATLDH ITNMIMRHGK KEKAQTILSR ALYLVYCQTR QDPIQALEKS LDELAPLMMT KTFNTGVAK ASVIPVPLNK RQRNRIAWNW IVQSANQRVS SDFAVRLGEE LTAIAKGTSS AFEKRDQIHK TAIAHRAYIQ L K UniProtKB: Small ribosomal subunit protein uS7m |
+Macromolecule #48: uS8m
| Macromolecule | Name: uS8m / type: protein_or_peptide / ID: 48 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 17.365404 KDa |
| Sequence | String: SLVKLANTCA HLQNCSKVRV ALTSIPYTKL QLQFAYNLYQ QGFLSSLQKG STMGPDKDFV EVTPDNISTR RLWVGLKYRD NKPVLSSCK LISKPNSRIH LPMEDMKKLC SGVTIRNIKP LQPGELILVR AHNNIMDINE AISKKLDGEV LCRVK UniProtKB: Small ribosomal subunit protein uS8m |
+Macromolecule #49: uS9m
| Macromolecule | Name: uS9m / type: protein_or_peptide / ID: 49 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 27.960496 KDa |
| Sequence | String: RRIVPKLATF YSANPNHEDR INRLERLLRK YIKLPSQNNN EAQQTKAPWI SFDEYALIGG GTKLKPTQYT QLLYMLNKLH NIDPQLTND EITSELSQYY KKSSMLSNNI KIKTLDEFGR SIAVGKRKSS TAKVFVVRGT GEILVNGRQL NDYFLKMKDR E SIMYPLQV ...String: RRIVPKLATF YSANPNHEDR INRLERLLRK YIKLPSQNNN EAQQTKAPWI SFDEYALIGG GTKLKPTQYT QLLYMLNKLH NIDPQLTND EITSELSQYY KKSSMLSNNI KIKTLDEFGR SIAVGKRKSS TAKVFVVRGT GEILVNGRQL NDYFLKMKDR E SIMYPLQV IESVGKYNIF ATTSGGGPTG QAESIMHAIA KALVVFNPLL KSRLHKAGVL TRDYRHVERK KPGKKKARKM PT WVKR UniProtKB: Small ribosomal subunit protein uS9m |
+Macromolecule #50: uS10m
| Macromolecule | Name: uS10m / type: protein_or_peptide / ID: 50 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 21.395377 KDa |
| Sequence | String: STRPYPVNVE AVYYAPLKLP IKYGDLVADI QLRSYDNENL DFYSDFILRT GYYLGIPLTG PKPLPTRRER WTVIKSPFVH AKSKENFER HTHKRLIRAW DTNPEVLQML IAYITKHSMA GVGMKCNFFQ RSEISLDLGS DANGLEKSLS NIDELYSLRN D DKAQTSAV GQKVLELLDS PDFKKHLE UniProtKB: Small ribosomal subunit protein uS10m |
+Macromolecule #51: uS11m
| Macromolecule | Name: uS11m / type: protein_or_peptide / ID: 51 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 16.716566 KDa |
| Sequence | String: KEEVVKYILH GKFTKNNTHL TFSSVVEDKN FHKNKGLTYN DTMLYYLNLP QKVKISLSTG CLGFRKAARG EYEAAFQTSG RMFELIKEK NMLNKDIEVV MDDFGKGRAA FISALVGKEG ASVVKKVVKI SDATKLKFGG VRSPKMRRL UniProtKB: Small ribosomal subunit protein uS11m |
+Macromolecule #52: uS12m
| Macromolecule | Name: uS12m / type: protein_or_peptide / ID: 52 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 13.5528 KDa |
| Sequence | String: ATLNQIKRGS GPPRRKKIST APQLDQCPQR KGVVLRVMVL KPKKPNSAQR KACRVRLTNG NVVSAYIPGE GHDAQEHSIV YVRGGRCQD LPGVKYHVIR GAGDLSGVVN RISSRSKYGA KKPSK UniProtKB: Small ribosomal subunit protein uS12m |
+Macromolecule #53: uS13m
| Macromolecule | Name: uS13m / type: protein_or_peptide / ID: 53 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 13.474945 KDa |
| Sequence | String: VVHILGKGFK GKEVIKIALA SKFYGIGKTT AEKICSKLGF YPWMRMHQLS EPQIMSIASE LSTMTIEGDA RAIVKDNIAL KRKIGSYSG MRHTLHLPVR GQHTRNNAKT ARKLNKIDRR G UniProtKB: Small ribosomal subunit protein uS13m |
+Macromolecule #54: uS14m
| Macromolecule | Name: uS14m / type: protein_or_peptide / ID: 54 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 13.563061 KDa |
| Sequence | String: MGNFRFPIKT KLPPGFINAR ILRDNFKRQQ FKENEILVKS LKFIARNMNL PTKLRLEAQL KLNALPNYMR STQIKNRCVD SGHARFVLS DFRLCRYQFR ENALKGNLPG VKKGIW UniProtKB: Small ribosomal subunit protein uS14m |
+Macromolecule #55: uS15m
| Macromolecule | Name: uS15m / type: protein_or_peptide / ID: 55 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 29.522064 KDa |
| Sequence | String: SAKAVKFLKA QRRKQKNEAK QATLKASTDK VDPVLGRADT PFITRIMAEL KEPLVLSKGY NIEEVDKFLA AIESAKRERA ELSGLNTEV VGIEDIEKLE DRREAILRIL SMRNSENKNA IKMAVELARK EFERFPGDTG SSEVQAACMT VRIQNMANHI K EHRKDFAN ...String: SAKAVKFLKA QRRKQKNEAK QATLKASTDK VDPVLGRADT PFITRIMAEL KEPLVLSKGY NIEEVDKFLA AIESAKRERA ELSGLNTEV VGIEDIEKLE DRREAILRIL SMRNSENKNA IKMAVELARK EFERFPGDTG SSEVQAACMT VRIQNMANHI K EHRKDFAN TRNLRILVQQ RQAILRYLKR DNPEKYYWTI QKLGLNDAAI TDEFNMDRRY MQDYEFFGDK ILIRDSKKVA NQ KRKEIRK QKRATF UniProtKB: Small ribosomal subunit protein uS15m |
+Macromolecule #56: bS16m
| Macromolecule | Name: bS16m / type: protein_or_peptide / ID: 56 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 13.402701 KDa |
| Sequence | String: TCGLVRIRLA RFGRKNSPVY NIVVANSRKA RDAKPIEVLG TYVPVPSPVT KRELKRGVVP IKDVKLDFDR TKYWIGVGAQ PSETVTKLL RKAGILNDAW ATSKNSNVNR KVVFERMETL UniProtKB: Small ribosomal subunit protein bS16m |
+Macromolecule #57: uS17m
| Macromolecule | Name: uS17m / type: protein_or_peptide / ID: 57 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 27.683986 KDa |
| Sequence | String: MARQNFLGLV VSQGKMQKTV KVRVETKVFN KKINKELFHR RDYLVHDEGE ISREGDLVRI EATRPLSKRK FFAIAEIIRN KGQQFALYE SEAQLSVAKE EAQKAKEFLD KRSVRENKLN EKTTLLRDIR TIQDALSSGS TPKELLEIKQ RYGIQDFSQE T VRQLLQLD ...String: MARQNFLGLV VSQGKMQKTV KVRVETKVFN KKINKELFHR RDYLVHDEGE ISREGDLVRI EATRPLSKRK FFAIAEIIRN KGQQFALYE SEAQLSVAKE EAQKAKEFLD KRSVRENKLN EKTTLLRDIR TIQDALSSGS TPKELLEIKQ RYGIQDFSQE T VRQLLQLD ISGLEVNLEK QRSLIDRIQT RLSELLSNDL KCDQFLKDHG VEDPLTLKKN IKKNLLRKHV MMDMQQPSQ UniProtKB: Small ribosomal subunit protein uS17m |
+Macromolecule #58: bS18m
| Macromolecule | Name: bS18m / type: protein_or_peptide / ID: 58 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 11.447264 KDa |
| Sequence | String: KIDQSLSKKL PKGTIYDPFD FSMGRIHLDR KYQANKNSNR NDIMKSGANP LEFYARPRIL SRYVTSTGRI QHRDITGLSA KNQRRLSKA IRRCQAIGLM UniProtKB: Small ribosomal subunit protein bS18m |
+Macromolecule #59: uS19m
| Macromolecule | Name: uS19m / type: protein_or_peptide / ID: 59 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 9.037476 KDa |
| Sequence | String: SRSVWKGPNI VPLPIREAMT KGTPIRTNAR AATILPQFVG LKFQIHNGKE YVPIEISEDM VGHKLGEFAP TRKRFSYTQT UniProtKB: Small ribosomal subunit protein uS19m |
+Macromolecule #60: bS21m
| Macromolecule | Name: bS21m / type: protein_or_peptide / ID: 60 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 10.872719 KDa |
| Sequence | String: AEIARSSVEN AQMRFNSGKS IIVNKNNPAE SFKRLNRIMF ENNIPGDKRS QRFYMKPGKV AELKRSQRHR KEFMMGFKRL IEIVKDAKR KGY UniProtKB: Small ribosomal subunit protein bS21m |
+Macromolecule #61: mS23
| Macromolecule | Name: mS23 / type: protein_or_peptide / ID: 61 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 27.101824 KDa |
| Sequence | String: MKIQTNAVNV LQRTSAYLKS GLLKETPAWY NVVASIPPST KFTREPRFKN PSNGHIIGKL VDVTEQPHAN NKGLYKTRPN SSDKRVGVK RLYRPPKLTY VEDRLRSLFY KQHPWELSRP KILVENEIGD ENYDWSHMLQ IGRPLDGESV IQRTMYLIKT K QYGDMVEA ...String: MKIQTNAVNV LQRTSAYLKS GLLKETPAWY NVVASIPPST KFTREPRFKN PSNGHIIGKL VDVTEQPHAN NKGLYKTRPN SSDKRVGVK RLYRPPKLTY VEDRLRSLFY KQHPWELSRP KILVENEIGD ENYDWSHMLQ IGRPLDGESV IQRTMYLIKT K QYGDMVEA YDHARYEFYA LRMQEETEQQ VALEEAEMFG SLFGVSAIEH GIQKEQEVLD VWEKKVVEET ELMAA UniProtKB: Small ribosomal subunit protein mS23 |
+Macromolecule #62: mS26
| Macromolecule | Name: mS26 / type: protein_or_peptide / ID: 62 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 26.737086 KDa |
| Sequence | String: GKGAAKYGFK SGVFPTTRSI LKSPTTKQTD IINKVKSPKP KGVLGIGYAK GVKHPKGSHR LSPKVNFIDV DNLIAKTVAE PQSIKSSNG SAQKVRLQKA ELRRKFLIEA FRKEEARLLH KHEYLQKRTK ELEKAKELEL EKLNKEKSSD LTIMTLDKMM S QPLLRNRS ...String: GKGAAKYGFK SGVFPTTRSI LKSPTTKQTD IINKVKSPKP KGVLGIGYAK GVKHPKGSHR LSPKVNFIDV DNLIAKTVAE PQSIKSSNG SAQKVRLQKA ELRRKFLIEA FRKEEARLLH KHEYLQKRTK ELEKAKELEL EKLNKEKSSD LTIMTLDKMM S QPLLRNRS PEESELLKLK RNYNRSLLNF QAHKKKLNEL LNLYHVANEF IVTESQLLKK IDKVFNDETE EFTDA UniProtKB: Small ribosomal subunit protein mS26 |
+Macromolecule #63: mS29
| Macromolecule | Name: mS29 / type: protein_or_peptide / ID: 63 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 45.650645 KDa |
| Sequence | String: RVTPGSLYKN WTNTTHTAQL QQTAVPLALP IFNFDDISKT LNKVVSYSNK QYKSLHHLGS FKKSQFNELF QKPVCLVRED ATNSFLKKL VSHPVKKFII TGEPGVGKTV LLSQAHAYAV DSKQIIINIS YPELFLNGRN DFSYDDDLKL FIQPMYLKKL I RKILKAND ...String: RVTPGSLYKN WTNTTHTAQL QQTAVPLALP IFNFDDISKT LNKVVSYSNK QYKSLHHLGS FKKSQFNELF QKPVCLVRED ATNSFLKKL VSHPVKKFII TGEPGVGKTV LLSQAHAYAV DSKQIIINIS YPELFLNGRN DFSYDDDLKL FIQPMYLKKL I RKILKAND PALLKSIELS KDYKFSNANP KNASVKPFVT LNKTKNTVLD LLSVMTHPHN RGKLMKAIID ELSVQSKVPI MF TVDNFSK VLTTAYSAYR NTENKQIYSL DLQMGKLMMD IISGETKFAN GESSTILAIS GVDRTNKTLP VALGKIPVDP YVT RYHYEP KFVELLQKGN VTEFEVPKLN KQEVNELIDY YKQSNVLLDK DITGKKWENL IDEKYFLSGN GNPRELLKSL VLSH R UniProtKB: Small ribosomal subunit protein mS29 |
+Macromolecule #64: mS33
| Macromolecule | Name: mS33 / type: protein_or_peptide / ID: 64 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 11.030979 KDa |
| Sequence | String: MNVPKARLLK VAELSAKIFD QNFNPSGIRT GSKILNERLK GPSVASYYGN PDILKFRHLK TLYPDIEFVD LEEQYRLSMV EAKKRRGKG APKKMKK UniProtKB: Small ribosomal subunit protein mS33 |
+Macromolecule #65: mS35
| Macromolecule | Name: mS35 / type: protein_or_peptide / ID: 65 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 32.334529 KDa |
| Sequence | String: SADLYMHPEK WKGLPPQRIL ELYWERMARL GSEYKPNKDE LNALLTTSEY SNVPVNDIKK LYHRGEQGAI DIKGGNVNRD NSLRPFMFD ELPSQAQELV AQHREQRFYN RLAAYELPLL AQYRQEYKRP SPESHPVTYR YTSYVGEEHP NSRKVVLSVK T KELGLEEK ...String: SADLYMHPEK WKGLPPQRIL ELYWERMARL GSEYKPNKDE LNALLTTSEY SNVPVNDIKK LYHRGEQGAI DIKGGNVNRD NSLRPFMFD ELPSQAQELV AQHREQRFYN RLAAYELPLL AQYRQEYKRP SPESHPVTYR YTSYVGEEHP NSRKVVLSVK T KELGLEEK SLHKFRILAR SRYDHTTDIF KMSSDKFEHA SQNARYLHDI LQRLLAESKD LTEDDFSDVP LDTRHTIAKS LR KKKRDYE FPEHWKRPED APKKKFDIVD QLLSTL UniProtKB: Small ribosomal subunit protein mS35 |
+Macromolecule #66: mS37
| Macromolecule | Name: mS37 / type: protein_or_peptide / ID: 66 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 10.305092 KDa |
| Sequence | String: PPVYRLPPLP RLKVKKPIIR QEANKCLVLM SNLLQCWSSY GHMSPKCAGL VTELKSCTSE SALGKRNNVQ KSNINYHAAR LYDRINGKP HD UniProtKB: Small ribosomal subunit protein mS37 |
+Macromolecule #67: mS38
| Macromolecule | Name: mS38 / type: protein_or_peptide / ID: 67 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 4.383451 KDa |
| Sequence | String: DSVMRKRKKK MKKHKLRKRR KREKAERRKL SQGR UniProtKB: Small ribosomal subunit protein mS38 |
+Macromolecule #68: mS41
| Macromolecule | Name: mS41 / type: protein_or_peptide / ID: 68 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 11.796482 KDa |
| Sequence | String: TIPKPSDQVP DVDAFLNKIG RNCNELKDTF ENNWNNLFQW DSKILKEKGV NIQQRKYILK QVHNYRNNRP IHEIKLGKKS FFGGERKRK AFTAKWKAEN UniProtKB: Small ribosomal subunit protein mS41 |
+Macromolecule #69: mS42
| Macromolecule | Name: mS42 / type: protein_or_peptide / ID: 69 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 29.036896 KDa |
| Sequence | String: IHVVPKLPNS KALLQNGVPN ILSSSGFKTV WFDYQRYLCD KLTLATAGQS LESYYPFHIL LKTAGNPLQS NIFNLASSIH NNHLFVENI LPSAVEHGTN SNAVVKTEPS RLFLSKIKDS FNGSDWEVVK EEMIYRAENE VLGQGWLFLV ENNEKKLFIL T SNNNGTPY ...String: IHVVPKLPNS KALLQNGVPN ILSSSGFKTV WFDYQRYLCD KLTLATAGQS LESYYPFHIL LKTAGNPLQS NIFNLASSIH NNHLFVENI LPSAVEHGTN SNAVVKTEPS RLFLSKIKDS FNGSDWEVVK EEMIYRAENE VLGQGWLFLV ENNEKKLFIL T SNNNGTPY YFPRNQSFDL NSAISIDEFA TLKQMKELIG KSTKLNGKVQ DWTMPIICVN LWDHAYLHDY GVGNRSKYVK NV LDNLNWS VVNNRIFS UniProtKB: Small ribosomal subunit protein mS42 |
+Macromolecule #70: mS43
| Macromolecule | Name: mS43 / type: protein_or_peptide / ID: 70 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 36.775781 KDa |
| Sequence | String: MLRFTGARAI RKYSTRYALE HLKEGAPLKG LFSIEGLQKA WFDRVKYLDA KLNDCTNEAQ QKPLETLIHE NSKSASKKHI VNYASSLYN LKFSMSSLQG CIRTPPEECP RLGPEALLQT PDFNRTISNE PLTTGNERLQ AALISSFGSL MEFRTLLINS N LAISGDGF ...String: MLRFTGARAI RKYSTRYALE HLKEGAPLKG LFSIEGLQKA WFDRVKYLDA KLNDCTNEAQ QKPLETLIHE NSKSASKKHI VNYASSLYN LKFSMSSLQG CIRTPPEECP RLGPEALLQT PDFNRTISNE PLTTGNERLQ AALISSFGSL MEFRTLLINS N LAISGDGF TWLVARRQLD KRAMRNDMPN RDIEYDKLFI LNTYNAGTPF NFSTSGVMNE LNNQYTNMEK QRAKEAGNLE DS EMTAKQA KTKFIYETQQ KGFSGKEVSY IPLLAIDASP KTWLTDYGVF GKREYLERVW DSIEWKIVES RLPQRTKIQA FNT L UniProtKB: Small ribosomal subunit protein mS43 |
+Macromolecule #71: mS44
| Macromolecule | Name: mS44 / type: protein_or_peptide / ID: 71 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 39.045031 KDa |
| Sequence | String: MGTITVVINE GPILLIRALH RATTNKKMFR STVWRRFAST GEIAKAKLDE FLIYHKTDAK LKPFIYRPKN AQILLTKDIR DPKTREPLQ PRPPVKPLSK QTLNDFIYSV EPNSTELLDW FKEWTGTSIR KRAIWTYISP IHVQKMLTAS FFKIGKYAHM V GLLYGIEH ...String: MGTITVVINE GPILLIRALH RATTNKKMFR STVWRRFAST GEIAKAKLDE FLIYHKTDAK LKPFIYRPKN AQILLTKDIR DPKTREPLQ PRPPVKPLSK QTLNDFIYSV EPNSTELLDW FKEWTGTSIR KRAIWTYISP IHVQKMLTAS FFKIGKYAHM V GLLYGIEH KFLKAQNPSV FDIEHFFNTN IMCALHRNRL KDYKDAEIAQ RKLQVAWKKV LNRKNNTGLA NILVATLGRQ IG FTPELTG LQPVDISLPD IPNSSSGAEL KDLLSKYEGI YLIARTLLDI DQHNAQYLEL QEFIRQYQNA LSESSDPYDT HLK ALGLLE TPPPQESTEK EEK UniProtKB: Small ribosomal subunit protein mS27 |
+Macromolecule #72: mS45
| Macromolecule | Name: mS45 / type: protein_or_peptide / ID: 72 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 36.763957 KDa |
| Sequence | String: FSRRRIAYPF YPFKKLGRQH PKKHDTNLKT AMRQFLGPKN YKGEYVMNKY FTVPTNHVPN YIKPDLERGQ SLEHPVTKKP LQLRYDGTL GPPPVENKRL QNIFKDRLLQ PFPSNPHCKT NYVLSPQLKQ SIFEEITVEG LSAQQVSQKY GLKIPRVEAI V KLVSVENS ...String: FSRRRIAYPF YPFKKLGRQH PKKHDTNLKT AMRQFLGPKN YKGEYVMNKY FTVPTNHVPN YIKPDLERGQ SLEHPVTKKP LQLRYDGTL GPPPVENKRL QNIFKDRLLQ PFPSNPHCKT NYVLSPQLKQ SIFEEITVEG LSAQQVSQKY GLKIPRVEAI V KLVSVENS WNRRNRVSSD LKTMDETLYR MFPVFDSDAS FKRENLSEIP VPQKTLASRF LTIAESEPFG PVDAAHVLEL EP AVETLRN LSTVGEHSSG HQQSTNKNTK VIYGELVEGE RSQYKFTNAK VGKVGYRYGS GNRDNKKDRR IGFNKLGQMV YI UniProtKB: Small ribosomal subunit protein mS45 |
+Macromolecule #73: mS46
| Macromolecule | Name: mS46 / type: protein_or_peptide / ID: 73 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 18.897553 KDa |
| Sequence | String: SSEYVLEEPT PLSLLEYTPQ VFPTKESRLV NFTLDSLKKS NYPIYRSPNL GILKVHDFTL NTPNFGKYTP GSSLIFAKEP QLQNLLIEE DPEDFHRQVT GEYQLLKPYV KKDFEKLTKS KDTVSKLVQN SQVVRLSLQS VVMGSEEKKL VYDVCSGMKP I SELQQ UniProtKB: Small ribosomal subunit protein mS46 |
+Macromolecule #74: mS47
| Macromolecule | Name: mS47 / type: protein_or_peptide / ID: 74 / Number of copies: 1 / Enantiomer: LEVO / EC number: 3-hydroxyisobutyryl-CoA hydrolase |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 51.367457 KDa |
| Sequence | String: APPVLFTVQD TARVITLNRP KKLNALNAEM SESMFKTLNE YAKSDTTNLV ILKSSNRPRS FCAGGDVATV AIFNFNKEFA KSIKFFTDE YSLNFQIATY LKPIVTFMDG ITMGGGVGLS IHTPFRIATE NTKWAMPEMD IGFFPDVGST FALPRIVTLA N SNSQMALY ...String: APPVLFTVQD TARVITLNRP KKLNALNAEM SESMFKTLNE YAKSDTTNLV ILKSSNRPRS FCAGGDVATV AIFNFNKEFA KSIKFFTDE YSLNFQIATY LKPIVTFMDG ITMGGGVGLS IHTPFRIATE NTKWAMPEMD IGFFPDVGST FALPRIVTLA N SNSQMALY LCLTGEVVTG ADAYMLGLAS HYVSSENLDA LQKRLGEISP PFNNDPQSAY FFGMVNESID EFVSPLPKDY VF KYSNEKL NVIEACFNLS KNGTIEDIMN NLRQYEGSAE GKAFAQEIKT KLLTKSPSSL QIALRLVQEN SRDHIESAIK RDL YTAANM CMNQDSLVEF SEATKHKLID KQRVPYPWTK KEQLFVSQLT SITSPKPSLP MSLLRNTSNV TWTQYPYHSK YQLP TEQEI AAYIEKRTND DTGAKVTERE VLNHFANVIP SRRGKLGIQS LCKIVCERKC EE UniProtKB: Small ribosomal subunit protein mS47 |
+Macromolecule #77: unknown protein sequence 1
| Macromolecule | Name: unknown protein sequence 1 / type: protein_or_peptide / ID: 77 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 8.017875 KDa |
| Sequence | String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) ...String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) |
+Macromolecule #78: unknown protein sequence 2
| Macromolecule | Name: unknown protein sequence 2 / type: protein_or_peptide / ID: 78 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 12.868854 KDa |
| Sequence | String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) ...String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) |
+Macromolecule #79: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 79 / Number of copies: 299 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
+Macromolecule #80: SODIUM ION
| Macromolecule | Name: SODIUM ION / type: ligand / ID: 80 / Number of copies: 1 |
|---|---|
| Molecular weight | Theoretical: 22.99 Da |
+Macromolecule #81: ZINC ION
| Macromolecule | Name: ZINC ION / type: ligand / ID: 81 / Number of copies: 2 / Formula: ZN |
|---|---|
| Molecular weight | Theoretical: 65.409 Da |
+Macromolecule #82: GUANOSINE-5'-DIPHOSPHATE
| Macromolecule | Name: GUANOSINE-5'-DIPHOSPHATE / type: ligand / ID: 82 / Number of copies: 1 / Formula: GDP |
|---|---|
| Molecular weight | Theoretical: 443.201 Da |
| Chemical component information |  ChemComp-GDP: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON II (4k x 4k) / Average electron dose: 23.5 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
| CTF correction | Software - Name: RELION (ver. 1.4) / Type: PHASE FLIPPING AND AMPLITUDE CORRECTION |
|---|---|
| Startup model | Type of model: EMDB MAP |
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.25 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: RELION (ver. 1.4) / Number images used: 141795 |
| Initial angle assignment | Type: OTHER / Software - Name: RELION (ver. 1.4) |
| Final angle assignment | Type: ANGULAR RECONSTITUTION / Software - Name: RELION (ver. 1.4) |
+ About Yorodumi
About Yorodumi
-News
-Feb 9, 2022. New format data for meta-information of EMDB entries
New format data for meta-information of EMDB entries
- Version 3 of the EMDB header file is now the official format.
- The previous official version 1.9 will be removed from the archive.
Related info.: EMDB header
EMDB header
External links: wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
-Aug 12, 2020. Covid-19 info
Covid-19 info

URL: https://pdbj.org/emnavi/covid19.php
New page: Covid-19 featured information page in EM Navigator.
Related info.: Covid-19 info /
Covid-19 info /  Mar 5, 2020. Novel coronavirus structure data
Mar 5, 2020. Novel coronavirus structure data
+Mar 5, 2020. Novel coronavirus structure data
Novel coronavirus structure data
- International Committee on Taxonomy of Viruses (ICTV) defined the short name of the 2019 coronavirus as "SARS-CoV-2".
- In the structure databanks used in Yorodumi, some data are registered as the other names, "COVID-19 virus" and "2019-nCoV". Here are the details of the virus and the list of structure data.
Related info.: Yorodumi Speices /
Yorodumi Speices /  Aug 12, 2020. Covid-19 info
Aug 12, 2020. Covid-19 info
External links: COVID-19 featured content - PDBj /
COVID-19 featured content - PDBj /  Molecule of the Month (242):Coronavirus Proteases
Molecule of the Month (242):Coronavirus Proteases
+Jan 31, 2019. EMDB accession codes are about to change! (news from PDBe EMDB page)
EMDB accession codes are about to change! (news from PDBe EMDB page)
- The allocation of 4 digits for EMDB accession codes will soon come to an end. Whilst these codes will remain in use, new EMDB accession codes will include an additional digit and will expand incrementally as the available range of codes is exhausted. The current 4-digit format prefixed with “EMD-” (i.e. EMD-XXXX) will advance to a 5-digit format (i.e. EMD-XXXXX), and so on. It is currently estimated that the 4-digit codes will be depleted around Spring 2019, at which point the 5-digit format will come into force.
- The EM Navigator/Yorodumi systems omit the EMD- prefix.
Related info.: Q: What is EMD? /
Q: What is EMD? /  ID/Accession-code notation in Yorodumi/EM Navigator
ID/Accession-code notation in Yorodumi/EM Navigator
External links: EMDB Accession Codes are Changing Soon! /
EMDB Accession Codes are Changing Soon! /  Contact to PDBj
Contact to PDBj
+Jul 12, 2017. Major update of PDB
Major update of PDB
- wwPDB released updated PDB data conforming to the new PDBx/mmCIF dictionary.
- This is a major update changing the version number from 4 to 5, and with Remediation, in which all the entries are updated.
- In this update, many items about electron microscopy experimental information are reorganized (e.g. em_software).
- Now, EM Navigator and Yorodumi are based on the updated data.
External links: wwPDB Remediation /
wwPDB Remediation /  Enriched Model Files Conforming to OneDep Data Standards Now Available in the PDB FTP Archive
Enriched Model Files Conforming to OneDep Data Standards Now Available in the PDB FTP Archive
-Yorodumi
Thousand views of thousand structures
- Yorodumi is a browser for structure data from EMDB, PDB, SASBDB, etc.
- This page is also the successor to EM Navigator detail page, and also detail information page/front-end page for Omokage search.
- The word "yorodu" (or yorozu) is an old Japanese word meaning "ten thousand". "mi" (miru) is to see.
Related info.: EMDB /
EMDB /  PDB /
PDB /  SASBDB /
SASBDB /  Comparison of 3 databanks /
Comparison of 3 databanks /  Yorodumi Search /
Yorodumi Search /  Aug 31, 2016. New EM Navigator & Yorodumi /
Aug 31, 2016. New EM Navigator & Yorodumi /  Yorodumi Papers /
Yorodumi Papers /  Jmol/JSmol /
Jmol/JSmol /  Function and homology information /
Function and homology information /  Changes in new EM Navigator and Yorodumi
Changes in new EM Navigator and Yorodumi
 Movie
Movie Controller
Controller























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)