[English] 日本語
 Yorodumi
Yorodumi- EMDB-35403: Paired helical filament from Alzheimer's disease brain with PET l... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Paired helical filament from Alzheimer's disease brain with PET ligand flortaucipir (+flortaucipir) | |||||||||||||||||||||
 Map data Map data | ||||||||||||||||||||||
 Sample Sample |
| |||||||||||||||||||||
 Keywords Keywords | Alzheimer's disease / Tau filaments / Paired helical filament / PET ligand / Flortaucipir / PROTEIN FIBRIL | |||||||||||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||||||||||||||
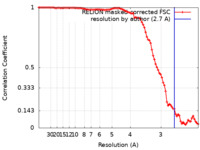
| Method | helical reconstruction / cryo EM / Resolution: 2.7 Å | |||||||||||||||||||||
 Authors Authors | Shi Y / Ghetti B / Goedert M / Scheres SHW | |||||||||||||||||||||
| Funding support |  United Kingdom, United Kingdom,  Switzerland, Switzerland,  United States, 6 items United States, 6 items
| |||||||||||||||||||||
 Citation Citation |  Journal: J Mol Biol / Year: 2023 Journal: J Mol Biol / Year: 2023Title: Cryo-EM Structures of Chronic Traumatic Encephalopathy Tau Filaments with PET Ligand Flortaucipir. Authors: Yang Shi / Bernardino Ghetti / Michel Goedert / Sjors H W Scheres /   Abstract: Positron emission tomography (PET) imaging allows monitoring the progression of amyloid aggregation in the living brain. [F]-Flortaucipir is the only approved PET tracer compound for the ...Positron emission tomography (PET) imaging allows monitoring the progression of amyloid aggregation in the living brain. [F]-Flortaucipir is the only approved PET tracer compound for the visualisation of tau aggregation. Here, we describe cryo-EM experiments on tau filaments in the presence and absence of flortaucipir. We used tau filaments isolated from the brain of an individual with Alzheimer's disease (AD), and from the brain of an individual with primary age-related tauopathy (PART) with a co-pathology of chronic traumatic encephalopathy (CTE). Unexpectedly, we were unable to visualise additional cryo-EM density for flortaucipir for AD paired helical or straight filaments (PHFs or SFs), but we did observe density for flortaucipir binding to CTE Type I filaments from the case with PART. In the latter, flortaucipir binds in a 1:1 molecular stoichiometry with tau, adjacent to lysine 353 and aspartate 358. By adopting a tilted geometry with respect to the helical axis, the 4.7 Å distance between neighbouring tau monomers is reconciled with the 3.5 Å distance consistent with π-π-stacking between neighbouring molecules of flortaucipir. | |||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_35403.map.gz emd_35403.map.gz | 24.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-35403-v30.xml emd-35403-v30.xml emd-35403.xml emd-35403.xml | 14.6 KB 14.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_35403_fsc.xml emd_35403_fsc.xml | 9.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_35403.png emd_35403.png | 40.8 KB | ||
| Masks |  emd_35403_msk_1.map emd_35403_msk_1.map | 64 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-35403.cif.gz emd-35403.cif.gz | 4.7 KB | ||
| Others |  emd_35403_half_map_1.map.gz emd_35403_half_map_1.map.gz emd_35403_half_map_2.map.gz emd_35403_half_map_2.map.gz | 49.3 MB 49.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-35403 http://ftp.pdbj.org/pub/emdb/structures/EMD-35403 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-35403 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-35403 | HTTPS FTP |
-Validation report
| Summary document |  emd_35403_validation.pdf.gz emd_35403_validation.pdf.gz | 911.3 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_35403_full_validation.pdf.gz emd_35403_full_validation.pdf.gz | 910.9 KB | Display | |
| Data in XML |  emd_35403_validation.xml.gz emd_35403_validation.xml.gz | 16.3 KB | Display | |
| Data in CIF |  emd_35403_validation.cif.gz emd_35403_validation.cif.gz | 21.2 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35403 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35403 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35403 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35403 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_35403.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_35403.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.15 Å | ||||||||||||||||||||||||||||||||||||
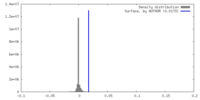
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
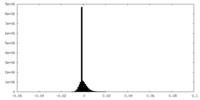
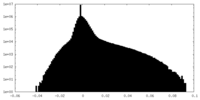
-Mask #1
| File |  emd_35403_msk_1.map emd_35403_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
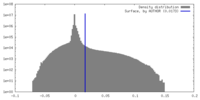
| Density Histograms |
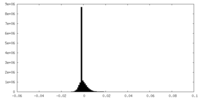
-Half map: #1
| File | emd_35403_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
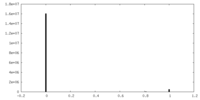
| Density Histograms |
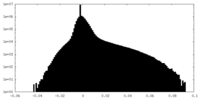
-Half map: #2
| File | emd_35403_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
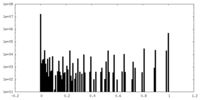
| Density Histograms |
- Sample components
Sample components
-Entire : Sarkosyl-insoluble fraction from the frontal cortex of an individ...
| Entire | Name: Sarkosyl-insoluble fraction from the frontal cortex of an individual with Alzheimer's disease |
|---|---|
| Components |
|
-Supramolecule #1: Sarkosyl-insoluble fraction from the frontal cortex of an individ...
| Supramolecule | Name: Sarkosyl-insoluble fraction from the frontal cortex of an individual with Alzheimer's disease type: tissue / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Microtubule-associated protein tau
| Macromolecule | Name: Microtubule-associated protein tau / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MAEPRQEFEV MEDHAGTYGL GDRKDQGGYT MHQDQEGDTD AGLKESPLQT PTEDGSEEPG SETSDAKSTP TAEDVTAPLV DEGAPGKQAA AQPHTEIPEG TTAEEAGIGD TPSLEDEAAG HVTQARMVSK SKDGTGSDDK KAKGADGKTK IATPRGAAPP GQKGQANATR ...String: MAEPRQEFEV MEDHAGTYGL GDRKDQGGYT MHQDQEGDTD AGLKESPLQT PTEDGSEEPG SETSDAKSTP TAEDVTAPLV DEGAPGKQAA AQPHTEIPEG TTAEEAGIGD TPSLEDEAAG HVTQARMVSK SKDGTGSDDK KAKGADGKTK IATPRGAAPP GQKGQANATR IPAKTPPAPK TPPSSGEPPK SGDRSGYSSP GSPGTPGSRS RTPSLPTPPT REPKKVAVVR TPPKSPSSAK SRLQTAPVPM PDLKNVKSKI GSTENLKHQP GGGKVQIINK KLDLSNVQSK CGSKDNIKHV PGGGSVQIVY KPVDLSKVTS KCGSLGNIHH KPGGGQVEVK SEKLDFKDRV QSKIGSLDNI THVPGGGNKK IETHKLTFRE NAKAKTDHGA EIVYKSPVVS GDTSPRHLSN VSSTGSIDMV DSPQLATLAD EVSASLAKQG L |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)