+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of Eaf3 CHD in complex with nucleosome | |||||||||
 Map data Map data | EM map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Histone modification binding domain / Histone deacetylase complex / GENE REGULATION | |||||||||
| Function / homology |  Function and homology information Function and homology informationRpd3S complex / NuA4 histone acetyltransferase complex / structural constituent of chromatin / nucleosome / nucleosome assembly / chromatin remodeling / protein heterodimerization activity / DNA repair / regulation of DNA-templated transcription / DNA binding / nucleus Similarity search - Function | |||||||||
| Biological species |   | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.4 Å | |||||||||
 Authors Authors | Cui H / Wang H | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2023 Journal: Nat Struct Mol Biol / Year: 2023Title: Structure of histone deacetylase complex Rpd3S bound to nucleosome. Authors: Wulong Li / Hengjun Cui / Zhimin Lu / Haibo Wang /  Abstract: Crosstalk between histone modifications represents a fundamental epigenetic mechanism in gene regulation. During the transcription elongation process, the histone deacetylase complex Rpd3S is ...Crosstalk between histone modifications represents a fundamental epigenetic mechanism in gene regulation. During the transcription elongation process, the histone deacetylase complex Rpd3S is recruited to H3K36-methylated nucleosomes to suppress cryptic transcription initiation. However, how subunits of Rpd3S are assembled and coordinated to recognize nucleosomal substrates and exert their deacetylation function remains unclear. Here we report the structure of Saccharomyces cerevisiae Rpd3S deacetylase bound to H3K36me3-modified nucleosome at 3.1 Å resolution. It shows that Sin3 and Rco1 subunits orchestrate the assembly of the complex and mediate its contact with nucleosome at multiple sites, with the Sin3-DNA interface as a pivotal anchor. The PHD1 domain of Rco1 recognizes the unmodified H3K4 and places the following H3 tail toward the active site of Rpd3, while the chromodomain of Eaf3 subunit recognizes the H3K36me3 mark and contacts both nucleosomal and linker DNA. The second copy of Eaf3-Rco1 is involved in neighboring nucleosome binding. Our work unravels the structural basis of chromatin targeting and deacetylation by the Rpd3S complex. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_35083.map.gz emd_35083.map.gz | 49.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-35083-v30.xml emd-35083-v30.xml emd-35083.xml emd-35083.xml | 23.1 KB 23.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_35083_fsc.xml emd_35083_fsc.xml | 9.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_35083.png emd_35083.png | 117.8 KB | ||
| Filedesc metadata |  emd-35083.cif.gz emd-35083.cif.gz | 7 KB | ||
| Others |  emd_35083_half_map_1.map.gz emd_35083_half_map_1.map.gz emd_35083_half_map_2.map.gz emd_35083_half_map_2.map.gz | 49.9 MB 49.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-35083 http://ftp.pdbj.org/pub/emdb/structures/EMD-35083 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-35083 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-35083 | HTTPS FTP |
-Validation report
| Summary document |  emd_35083_validation.pdf.gz emd_35083_validation.pdf.gz | 860.7 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_35083_full_validation.pdf.gz emd_35083_full_validation.pdf.gz | 860.3 KB | Display | |
| Data in XML |  emd_35083_validation.xml.gz emd_35083_validation.xml.gz | 16.2 KB | Display | |
| Data in CIF |  emd_35083_validation.cif.gz emd_35083_validation.cif.gz | 21.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35083 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35083 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35083 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35083 | HTTPS FTP |
-Related structure data
| Related structure data |  8hxzMC  8hxxC  8hxyC  8hy0C  8jhoC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_35083.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_35083.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | EM map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.05 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: EM half1 map
| File | emd_35083_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | EM half1 map | ||||||||||||
| Projections & Slices |
| ||||||||||||
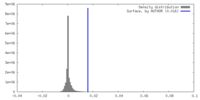
| Density Histograms |
-Half map: EM half2 map
| File | emd_35083_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | EM half2 map | ||||||||||||
| Projections & Slices |
| ||||||||||||
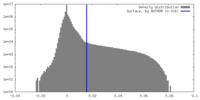
| Density Histograms |
- Sample components
Sample components
-Entire : Eaf3 CHD in complex with nucleosome
| Entire | Name: Eaf3 CHD in complex with nucleosome |
|---|---|
| Components |
|
-Supramolecule #1: Eaf3 CHD in complex with nucleosome
| Supramolecule | Name: Eaf3 CHD in complex with nucleosome / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#7 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 150 KDa |
-Macromolecule #1: Histone H3
| Macromolecule | Name: Histone H3 / type: protein_or_peptide / ID: 1 Details: Cys110 residues of chain A/E were mutated to Ala due to the preparation of ML3-modified nucleosome. Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: |
| Molecular weight | Theoretical: 15.331982 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: ARTKQTARKS TGGKAPRKQL ATKAARKSAP ATGGV(ML3)KPHR YRPGTVALRE IRRYQKSTEL LIRKLPFQRL VREIAQ DFK TDLRFQSSAV MALQEASEAY LVALFEDTNL AAIHAKRVTI MPKDIQLARR IRGERA UniProtKB: Histone H3 |
-Macromolecule #2: Histone H4
| Macromolecule | Name: Histone H4 / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: |
| Molecular weight | Theoretical: 11.263231 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: SGRGKGGKGL GKGGAKRHRK VLRDNIQGIT KPAIRRLARR GGVKRISGLI YEETRGVLKV FLENVIRDAV TYTEHAKRKT VTAMDVVYA LKRQGRTLYG FGG UniProtKB: Histone H4 |
-Macromolecule #3: Histone H2A
| Macromolecule | Name: Histone H2A / type: protein_or_peptide / ID: 3 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: |
| Molecular weight | Theoretical: 13.978241 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: SGRGKQGGKT RAKAKTRSSR AGLQFPVGRV HRLLRKGNYA ERVGAGAPVY LAAVLEYLTA EILELAGNAA RDNKKTRIIP RHLQLAVRN DEELNKLLGR VTIAQGGVLP NIQSVLLPKK TESSKSAKSK UniProtKB: Histone H2A |
-Macromolecule #4: Histone H2B
| Macromolecule | Name: Histone H2B / type: protein_or_peptide / ID: 4 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: |
| Molecular weight | Theoretical: 13.524752 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: AKSAPAPKKG SKKAVTKTQK KDGKKRRKTR KESYAIYVYK VLKQVHPDTG ISSKAMSIMN SFVNDVFERI AGEASRLAHY NKRSTITSR EIQTAVRLLL PGELAKHAVS EGTKAVTKYT SAK UniProtKB: Histone H2B |
-Macromolecule #7: Chromatin modification-related protein EAF3
| Macromolecule | Name: Chromatin modification-related protein EAF3 / type: protein_or_peptide / ID: 7 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 45.266406 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MVDLEQEFAL GGRCLAFHGP LMYEAKILKI WDPSSKMYTS IPNDKPGGSS QATKEIKPQK LGEDESIPEE IINGKCFFIH YQGWKSSWD EWVGYDRIRA YNEENIAMKK RLANEAKEAK KSLLEQQKKK KLSTSLGGPS NGGKRKGDSR SNASISKSTS Q SFLTSSVS ...String: MVDLEQEFAL GGRCLAFHGP LMYEAKILKI WDPSSKMYTS IPNDKPGGSS QATKEIKPQK LGEDESIPEE IINGKCFFIH YQGWKSSWD EWVGYDRIRA YNEENIAMKK RLANEAKEAK KSLLEQQKKK KLSTSLGGPS NGGKRKGDSR SNASISKSTS Q SFLTSSVS GRKSGRSSAN SLHPGSSLRS SSDQNGNDDR RRSSSLSPNM LHHIAGYPTP KISLQIPIKL KSVLVDDWEY VT KDKKICR LPADVTVEMV LNKYEHEVSQ ELESPGSQSQ LSEYCAGLKL YFDKCLGNML LYRLERLQYD ELLKKSSKDQ KPL VPIRIY GAIHLLRLIS VLPELISSTT MDLQSCQLLI KQTEDFLVWL LMHVDEYFND KDPNRSDDAL YVNTSSQYEG VALG M UniProtKB: Chromatin modification-related protein EAF3 |
-Macromolecule #5: DNA (352-MER)
| Macromolecule | Name: DNA (352-MER) / type: dna / ID: 5 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 108.340836 KDa |
| Sequence | String: (DG)(DA)(DA)(DT)(DT)(DC)(DG)(DA)(DT)(DA) (DT)(DC)(DG)(DA)(DG)(DA)(DA)(DT)(DC)(DC) (DC)(DG)(DG)(DT)(DG)(DC)(DC)(DG)(DA) (DG)(DG)(DC)(DC)(DG)(DC)(DT)(DC)(DA)(DA) (DT) (DT)(DG)(DG)(DT)(DC)(DG) ...String: (DG)(DA)(DA)(DT)(DT)(DC)(DG)(DA)(DT)(DA) (DT)(DC)(DG)(DA)(DG)(DA)(DA)(DT)(DC)(DC) (DC)(DG)(DG)(DT)(DG)(DC)(DC)(DG)(DA) (DG)(DG)(DC)(DC)(DG)(DC)(DT)(DC)(DA)(DA) (DT) (DT)(DG)(DG)(DT)(DC)(DG)(DT)(DA) (DG)(DA)(DC)(DA)(DG)(DC)(DT)(DC)(DT)(DA) (DG)(DC) (DA)(DC)(DC)(DG)(DC)(DT)(DT) (DA)(DA)(DA)(DC)(DG)(DC)(DA)(DC)(DG)(DT) (DA)(DC)(DG) (DC)(DG)(DC)(DT)(DG)(DT) (DC)(DC)(DC)(DC)(DC)(DG)(DC)(DG)(DT)(DT) (DT)(DT)(DA)(DA) (DC)(DC)(DG)(DC)(DC) (DA)(DA)(DG)(DG)(DG)(DG)(DA)(DT)(DT)(DA) (DC)(DT)(DC)(DC)(DC) (DT)(DA)(DG)(DT) (DC)(DT)(DC)(DC)(DA)(DG)(DG)(DC)(DA)(DC) (DG)(DT)(DG)(DT)(DC)(DA) (DG)(DA)(DT) (DA)(DT)(DA)(DT)(DA)(DC)(DA)(DT)(DC)(DC) (DT)(DG)(DT)(DG)(DC)(DA)(DT) (DG)(DT) (DA)(DT)(DT)(DG)(DA)(DA)(DA)(DG)(DT)(DA) (DC)(DT)(DG)(DC)(DC)(DA)(DG)(DT) (DT) (DC)(DT)(DA)(DG)(DA)(DC)(DT)(DG)(DG)(DA) (DG)(DA)(DA)(DT)(DC)(DC)(DC)(DG)(DG) (DT)(DG)(DC)(DC)(DG)(DA)(DG)(DG)(DC)(DC) (DG)(DC)(DT)(DC)(DA)(DA)(DT)(DT)(DG)(DG) (DT)(DC)(DG)(DT)(DA)(DG)(DA)(DC)(DA) (DG)(DC)(DT)(DC)(DT)(DA)(DG)(DC)(DA)(DC) (DC) (DG)(DC)(DT)(DT)(DA)(DA)(DA)(DC) (DG)(DC)(DA)(DC)(DG)(DT)(DA)(DC)(DG)(DC) (DG)(DC) (DT)(DG)(DT)(DC)(DC)(DC)(DC) (DC)(DG)(DC)(DG)(DT)(DT)(DT)(DT)(DA)(DA) (DC)(DC)(DG) (DC)(DC)(DA)(DA)(DG)(DG) (DG)(DG)(DA)(DT)(DT)(DA)(DC)(DT)(DC)(DC) (DC)(DT)(DA)(DG) (DT)(DC)(DT)(DC)(DC) (DA)(DG)(DG)(DC)(DA)(DC)(DG)(DT)(DG)(DT) (DC)(DA)(DG)(DA)(DT) (DA)(DT)(DA)(DT) (DA)(DC)(DA)(DT)(DC)(DC)(DT)(DG)(DT)(DG) (DC)(DA)(DT)(DG)(DT)(DA) (DT)(DT)(DG) (DA)(DA)(DC)(DA)(DG)(DC)(DG)(DA)(DT) |
-Macromolecule #6: DNA (352-MER)
| Macromolecule | Name: DNA (352-MER) / type: dna / ID: 6 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 109.079289 KDa |
| Sequence | String: (DA)(DT)(DC)(DG)(DC)(DT)(DG)(DT)(DT)(DC) (DA)(DA)(DT)(DA)(DC)(DA)(DT)(DG)(DC)(DA) (DC)(DA)(DG)(DG)(DA)(DT)(DG)(DT)(DA) (DT)(DA)(DT)(DA)(DT)(DC)(DT)(DG)(DA)(DC) (DA) (DC)(DG)(DT)(DG)(DC)(DC) ...String: (DA)(DT)(DC)(DG)(DC)(DT)(DG)(DT)(DT)(DC) (DA)(DA)(DT)(DA)(DC)(DA)(DT)(DG)(DC)(DA) (DC)(DA)(DG)(DG)(DA)(DT)(DG)(DT)(DA) (DT)(DA)(DT)(DA)(DT)(DC)(DT)(DG)(DA)(DC) (DA) (DC)(DG)(DT)(DG)(DC)(DC)(DT)(DG) (DG)(DA)(DG)(DA)(DC)(DT)(DA)(DG)(DG)(DG) (DA)(DG) (DT)(DA)(DA)(DT)(DC)(DC)(DC) (DC)(DT)(DT)(DG)(DG)(DC)(DG)(DG)(DT)(DT) (DA)(DA)(DA) (DA)(DC)(DG)(DC)(DG)(DG) (DG)(DG)(DG)(DA)(DC)(DA)(DG)(DC)(DG)(DC) (DG)(DT)(DA)(DC) (DG)(DT)(DG)(DC)(DG) (DT)(DT)(DT)(DA)(DA)(DG)(DC)(DG)(DG)(DT) (DG)(DC)(DT)(DA)(DG) (DA)(DG)(DC)(DT) (DG)(DT)(DC)(DT)(DA)(DC)(DG)(DA)(DC)(DC) (DA)(DA)(DT)(DT)(DG)(DA) (DG)(DC)(DG) (DG)(DC)(DC)(DT)(DC)(DG)(DG)(DC)(DA)(DC) (DC)(DG)(DG)(DG)(DA)(DT)(DT) (DC)(DT) (DC)(DC)(DA)(DG)(DT)(DC)(DT)(DA)(DG)(DA) (DA)(DC)(DT)(DG)(DG)(DC)(DA)(DG) (DT) (DA)(DC)(DT)(DT)(DT)(DC)(DA)(DA)(DT)(DA) (DC)(DA)(DT)(DG)(DC)(DA)(DC)(DA)(DG) (DG)(DA)(DT)(DG)(DT)(DA)(DT)(DA)(DT)(DA) (DT)(DC)(DT)(DG)(DA)(DC)(DA)(DC)(DG)(DT) (DG)(DC)(DC)(DT)(DG)(DG)(DA)(DG)(DA) (DC)(DT)(DA)(DG)(DG)(DG)(DA)(DG)(DT)(DA) (DA) (DT)(DC)(DC)(DC)(DC)(DT)(DT)(DG) (DG)(DC)(DG)(DG)(DT)(DT)(DA)(DA)(DA)(DA) (DC)(DG) (DC)(DG)(DG)(DG)(DG)(DG)(DA) (DC)(DA)(DG)(DC)(DG)(DC)(DG)(DT)(DA)(DC) (DG)(DT)(DG) (DC)(DG)(DT)(DT)(DT)(DA) (DA)(DG)(DC)(DG)(DG)(DT)(DG)(DC)(DT)(DA) (DG)(DA)(DG)(DC) (DT)(DG)(DT)(DC)(DT) (DA)(DC)(DG)(DA)(DC)(DC)(DA)(DA)(DT)(DT) (DG)(DA)(DG)(DC)(DG) (DG)(DC)(DC)(DT) (DC)(DG)(DG)(DC)(DA)(DC)(DC)(DG)(DG)(DG) (DA)(DT)(DT)(DC)(DT)(DC) (DG)(DA)(DT) (DA)(DT)(DC)(DG)(DA)(DA)(DT)(DT)(DC) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 Details: 20 mM HEPES-Na pH 7.5, 40 mM KCl, 2 mM MgCl2, 1 mM TCEP |
|---|---|
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277.15 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 44.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.8 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
|---|---|
| Output model |  PDB-8hxz: |
 Movie
Movie Controller
Controller















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)