[English] 日本語
 Yorodumi
Yorodumi- EMDB-34945: Cryo-EM structure of SARS-CoV-2 Omicron Prototype S-trimer in com... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of SARS-CoV-2 Omicron Prototype S-trimer in complex with fab L4.65 and L5.34 | |||||||||
 Map data Map data | Cryo-EM structure of SARS-CoV-2 Prototype S-trimer in complex with fab L4.65 and L5.34 | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | SARS-CoV-2 / Omicron Prototype S-trimer / Cryo-EM structure / fab / IMMUNE SYSTEM/VIRAL PROTEIN / IMMUNE SYSTEM-VIRAL PROTEIN complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationsymbiont-mediated disruption of host tissue / Maturation of spike protein / Translation of Structural Proteins / Virion Assembly and Release / host cell surface / host extracellular space / symbiont-mediated-mediated suppression of host tetherin activity / Induction of Cell-Cell Fusion / structural constituent of virion / membrane fusion ...symbiont-mediated disruption of host tissue / Maturation of spike protein / Translation of Structural Proteins / Virion Assembly and Release / host cell surface / host extracellular space / symbiont-mediated-mediated suppression of host tetherin activity / Induction of Cell-Cell Fusion / structural constituent of virion / membrane fusion / entry receptor-mediated virion attachment to host cell / Attachment and Entry / host cell endoplasmic reticulum-Golgi intermediate compartment membrane / positive regulation of viral entry into host cell / receptor-mediated virion attachment to host cell / host cell surface receptor binding / symbiont-mediated suppression of host innate immune response / endocytosis involved in viral entry into host cell / receptor ligand activity / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / symbiont entry into host cell / virion attachment to host cell / SARS-CoV-2 activates/modulates innate and adaptive immune responses / host cell plasma membrane / virion membrane / identical protein binding / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.65 Å | |||||||||
 Authors Authors | Gao GF / Liu S | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Cell Discov / Year: 2023 Journal: Cell Discov / Year: 2023Title: Dosing interval regimen shapes potency and breadth of antibody repertoire after vaccination of SARS-CoV-2 RBD protein subunit vaccine. Authors: Shuxin Guo / Yuxuan Zheng / Zhengrong Gao / Minrun Duan / Sheng Liu / Pan Du / XiaoYu Xu / Kun Xu / Xin Zhao / Yan Chai / Peiyi Wang / Qi Zhao / George F Gao / Lianpan Dai /  Abstract: Vaccination with different vaccines has been implemented globally to counter the continuous COVID-19 pandemic. However, the vaccine-elicited antibodies have reduced efficiency against the highly ...Vaccination with different vaccines has been implemented globally to counter the continuous COVID-19 pandemic. However, the vaccine-elicited antibodies have reduced efficiency against the highly mutated Omicron sub-variants. Previously, we developed a protein subunit COVID-19 vaccine called ZF2001, based on the dimeric receptor-binding domain (RBD). This vaccine has been administered using different dosing intervals in real-world setting. Some individuals received three doses of ZF2001, with a one-month interval between each dose, due to urgent clinical requirements. Others had an extended dosing interval of up to five months between the second and third dose, a standard vaccination regimen for the protein subunit vaccine against hepatitis B. In this study, we profile B cell responses in individuals who received three doses of ZF2001, and compared those with long or short dosing intervals. We observed that the long-interval group exhibited higher and broader serologic antibody responses. These responses were associated with the increased size and evolution of vaccine-elicited B-cell receptor repertoires, characterized by the elevation of expanded clonotypes and somatic hypermutations. Both groups of individuals generated substantial amounts of broadly neutralizing antibodies (bnAbs) against various SARS-CoV-2 variants, including Omicron sub-variants such as XBB. These bnAbs target four antigenic sites within the RBD. To determine the vulnerable site of SARS-CoV-2, we employed cryo-electron microscopy to identify the epitopes of highly potent bnAbs that targeted two major sites. Our findings provide immunological insights into the B cell responses elicited by RBD-based vaccine, and suggest that a vaccination regimen of prolonging time interval should be used in practice. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_34945.map.gz emd_34945.map.gz | 483.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-34945-v30.xml emd-34945-v30.xml emd-34945.xml emd-34945.xml | 22.6 KB 22.6 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_34945.png emd_34945.png | 112.3 KB | ||
| Filedesc metadata |  emd-34945.cif.gz emd-34945.cif.gz | 7.2 KB | ||
| Others |  emd_34945_half_map_1.map.gz emd_34945_half_map_1.map.gz emd_34945_half_map_2.map.gz emd_34945_half_map_2.map.gz | 475.6 MB 475.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-34945 http://ftp.pdbj.org/pub/emdb/structures/EMD-34945 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34945 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34945 | HTTPS FTP |
-Related structure data
| Related structure data |  8hpvMC  8hp9C  8hpfC  8hpqC  8hpuC  8hq7C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_34945.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_34945.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-EM structure of SARS-CoV-2 Prototype S-trimer in complex with fab L4.65 and L5.34 | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.856 Å | ||||||||||||||||||||||||||||||||||||
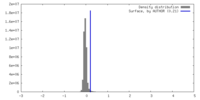
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Cryo-EM structure of SARS-CoV-2 Prototype S-trimer in complex...
| File | emd_34945_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-EM structure of SARS-CoV-2 Prototype S-trimer in complex with fab L4.65 and L5.34 | ||||||||||||
| Projections & Slices |
| ||||||||||||
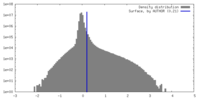
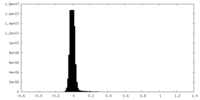
| Density Histograms |
-Half map: Cryo-EM structure of SARS-CoV-2 Prototype S-trimer in complex...
| File | emd_34945_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-EM structure of SARS-CoV-2 Prototype S-trimer in complex with fab L4.65 and L5.34 | ||||||||||||
| Projections & Slices |
| ||||||||||||
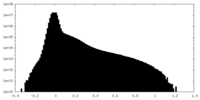
| Density Histograms |
- Sample components
Sample components
-Entire : Cryo-EM structure of SARS-CoV-2 Omicron Prototype S-trimer in com...
| Entire | Name: Cryo-EM structure of SARS-CoV-2 Omicron Prototype S-trimer in complex with fab L4.65 and L5.34 |
|---|---|
| Components |
|
-Supramolecule #1: Cryo-EM structure of SARS-CoV-2 Omicron Prototype S-trimer in com...
| Supramolecule | Name: Cryo-EM structure of SARS-CoV-2 Omicron Prototype S-trimer in complex with fab L4.65 and L5.34 type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Spike protein S2'
| Macromolecule | Name: Spike protein S2' / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 125.606203 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QCVNLTTRTQ LPPAYTNSFT RGVYYPDKVF RSSVLHSTQD LFLPFFSNVT WFHAIHVSGT NGTKRFDNPV LPFNDGVYFA STEKSNIIR GWIFGTTLDS KTQSLLIVNN ATNVVIKVCE FQFCNDPFLG VYYHKNNKSW MESEFRVYSS ANNCTFEYVS Q PFLMDLEG ...String: QCVNLTTRTQ LPPAYTNSFT RGVYYPDKVF RSSVLHSTQD LFLPFFSNVT WFHAIHVSGT NGTKRFDNPV LPFNDGVYFA STEKSNIIR GWIFGTTLDS KTQSLLIVNN ATNVVIKVCE FQFCNDPFLG VYYHKNNKSW MESEFRVYSS ANNCTFEYVS Q PFLMDLEG KQGNFKNLRE FVFKNIDGYF KIYSKHTPIN LVRDLPQGFS ALEPLVDLPI GINITRFQTL LALHRSYLTP GD SSSGWTA GAAAYYVGYL QPRTFLLKYN ENGTITDAVD CALDPLSETK CTLKSFTVEK GIYQTSNFRV QPTESIVRFP NIT NLCPFG EVFNATRFAS VYAWNRKRIS NCVADYSVLY NSASFSTFKC YGVSPTKLND LCFTNVYADS FVIRGDEVRQ IAPG QTGKI ADYNYKLPDD FTGCVIAWNS NNLDSKVGGN YNYLYRLFRK SNLKPFERDI STEIYQAGST PCNGVEGFNC YFPLQ SYGF QPTNGVGYQP YRVVVLSFEL LHAPATVCGP KKSTNLVKNK CVNFNFNGLT GTGVLTESNK KFLPFQQFGR DIADTT DAV RDPQTLEILD ITPCSFGGVS VITPGTNTSN QVAVLYQDVN CTEVPVAIHA DQLTPTWRVY STGSNVFQTR AGCLIGA EH VNNSYECDIP IGAGICASYQ TQTNSPRRAR SVASQSIIAY TMSLGAENSV AYSNNSIAIP TNFTISVTTE ILPVSMTK T SVDCTMYICG DSTECSNLLL QYGSFCTQLN RALTGIAVEQ DKNTQEVFAQ VKQIYKTPPI KDFGGFNFSQ ILPDPSKPS KRSPIEDLLF NKVTLADAGF IKQYGDCLGD IAARDLICAQ KFNGLTVLPP LLTDEMIAQY TSALLAGTIT SGWTFGAGPA LQIPFPMQM AYRFNGIGVT QNVLYENQKL IANQFNSAIG KIQDSLSSTP SALGKLQDVV NQNAQALNTL VKQLSSNFGA I SSVLNDIL SRLDPPEAEV QIDRLITGRL QSLQTYVTQQ LIRAAEIRAS ANLAATKMSE CVLGQSKRVD FCGKGYHLMS FP QSAPHGV VFLHVTYVPA QEKNFTTAPA ICHDGKAHFP REGVFVSNGT HWFVTQRNFY EPQIITTDNT FVSGNCDVVI GIV NNTVYD PLQPELDS UniProtKB: Spike glycoprotein |
-Macromolecule #2: fab L5.34
| Macromolecule | Name: fab L5.34 / type: protein_or_peptide / ID: 2 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 23.721539 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: EVQLVESGGG LVQPGGSLRL SCAASEITVS SNYMNWVRQA PGKGLEWVSV VYPGGSTFYT DSVKGRFTIS RDNSKNTLYL QMNSLRAED TAVYYCARES GGFPLAEGAF DIWGQGTMVT VSSASTKGPS VFPLAPSSKS TSGGTAALGC LVKDYFPEPV T VSWNSGAL ...String: EVQLVESGGG LVQPGGSLRL SCAASEITVS SNYMNWVRQA PGKGLEWVSV VYPGGSTFYT DSVKGRFTIS RDNSKNTLYL QMNSLRAED TAVYYCARES GGFPLAEGAF DIWGQGTMVT VSSASTKGPS VFPLAPSSKS TSGGTAALGC LVKDYFPEPV T VSWNSGAL TSGVHTFPAV LQSSGLYSLS SVVTVPSSSL GTQTYICNVN HKPSNTKVDK RVEPKSC |
-Macromolecule #3: fab L5.34
| Macromolecule | Name: fab L5.34 / type: protein_or_peptide / ID: 3 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 23.379928 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: DIQMTQSPSS LSASVGDRVS ITCRASQSIS THLHWYQQKP GKAPKLLISA ASTLQSGVPS RFSGSGSGTD FTLTITSLQP EDFATYYCQ QSYSTPRGLS FGGGTKVEIK RTVAAPSVFI FPPSDEQLKS GTASVVCLLN NFYPREAKVQ WKVDNALQSG N SQESVTEQ ...String: DIQMTQSPSS LSASVGDRVS ITCRASQSIS THLHWYQQKP GKAPKLLISA ASTLQSGVPS RFSGSGSGTD FTLTITSLQP EDFATYYCQ QSYSTPRGLS FGGGTKVEIK RTVAAPSVFI FPPSDEQLKS GTASVVCLLN NFYPREAKVQ WKVDNALQSG N SQESVTEQ DSKDSTYSLS STLTLSKADY EKHKVYACEV THQGLSSPVT KSFNRGEC |
-Macromolecule #4: fab L4.65
| Macromolecule | Name: fab L4.65 / type: protein_or_peptide / ID: 4 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 24.953938 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QITLKESGPT LVKPTQTLTL TCTFSGFSLS TSGVGVAWIR QPPGKALEWL ALIYWDNDKR SSPSLNNRLT ITKDTSKNQV VLTMTNMDP EDTATYYCAH FFSHYDSSNY YYGSWFDPWG QGTLVTVSSA STKGPSVFPL APSSKSTSGG TAALGCLVKD Y FPEPVTVS ...String: QITLKESGPT LVKPTQTLTL TCTFSGFSLS TSGVGVAWIR QPPGKALEWL ALIYWDNDKR SSPSLNNRLT ITKDTSKNQV VLTMTNMDP EDTATYYCAH FFSHYDSSNY YYGSWFDPWG QGTLVTVSSA STKGPSVFPL APSSKSTSGG TAALGCLVKD Y FPEPVTVS WNSGALTSGV HTFPAVLQSS GLYSLSSVVT VPSSSLGTQT YICNVNHKPS NTKVDKRVEP KSC |
-Macromolecule #5: fab L4.65
| Macromolecule | Name: fab L4.65 / type: protein_or_peptide / ID: 5 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 23.556061 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: EIVLTQSPGT LSLSPGERAT LSCRASQSFD SRYLGWYQQK SGQAPRLLIY GASSRATGIP DRFSGSGSGT DFTLTISRLE PEDFAVYYC QQFGDSPFTF GQGTKLEIKR TVAAPSVFIF PPSDEQLKSG TASVVCLLNN FYPREAKVQW KVDNALQSGN S QESVTEQD ...String: EIVLTQSPGT LSLSPGERAT LSCRASQSFD SRYLGWYQQK SGQAPRLLIY GASSRATGIP DRFSGSGSGT DFTLTISRLE PEDFAVYYC QQFGDSPFTF GQGTKLEIKR TVAAPSVFIF PPSDEQLKSG TASVVCLLNN FYPREAKVQW KVDNALQSGN S QESVTEQD SKDSTYSLSS TLTLSKADYE KHKVYACEVT HQGLSSPVTK SFNRGEC |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Average electron dose: 1.39 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 5.0 µm / Nominal defocus min: 1.2 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)