[English] 日本語
 Yorodumi
Yorodumi- EMDB-34944: Cryo-EM structure of SARS-CoV-2 Omicron BA.4 RBD in complex with ... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of SARS-CoV-2 Omicron BA.4 RBD in complex with fab L4.65 and L5.34 | |||||||||
 Map data Map data | Cryo-EM structure of SARS-CoV-2 Omicron BA.4 RBD in complex with fab L4.65 and L5.34 | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | SARS-CoV-2 / Omicron BA.4 RBD / Cryo-EM structure / fab / IMMUNE SYSTEM/VIRAL PROTEIN / IMMUNE SYSTEM-VIRAL PROTEIN complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationsymbiont-mediated disruption of host tissue / Maturation of spike protein / Translation of Structural Proteins / Virion Assembly and Release / host cell surface / host extracellular space / viral translation / symbiont-mediated-mediated suppression of host tetherin activity / Induction of Cell-Cell Fusion / structural constituent of virion ...symbiont-mediated disruption of host tissue / Maturation of spike protein / Translation of Structural Proteins / Virion Assembly and Release / host cell surface / host extracellular space / viral translation / symbiont-mediated-mediated suppression of host tetherin activity / Induction of Cell-Cell Fusion / structural constituent of virion / membrane fusion / entry receptor-mediated virion attachment to host cell / Attachment and Entry / host cell endoplasmic reticulum-Golgi intermediate compartment membrane / positive regulation of viral entry into host cell / receptor-mediated virion attachment to host cell / host cell surface receptor binding / symbiont-mediated suppression of host innate immune response / receptor ligand activity / endocytosis involved in viral entry into host cell / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / symbiont entry into host cell / virion attachment to host cell / SARS-CoV-2 activates/modulates innate and adaptive immune responses / host cell plasma membrane / virion membrane / identical protein binding / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.56 Å | |||||||||
 Authors Authors | Gao GF / Liu S | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Cell Discov / Year: 2023 Journal: Cell Discov / Year: 2023Title: Dosing interval regimen shapes potency and breadth of antibody repertoire after vaccination of SARS-CoV-2 RBD protein subunit vaccine. Authors: Shuxin Guo / Yuxuan Zheng / Zhengrong Gao / Minrun Duan / Sheng Liu / Pan Du / XiaoYu Xu / Kun Xu / Xin Zhao / Yan Chai / Peiyi Wang / Qi Zhao / George F Gao / Lianpan Dai /  Abstract: Vaccination with different vaccines has been implemented globally to counter the continuous COVID-19 pandemic. However, the vaccine-elicited antibodies have reduced efficiency against the highly ...Vaccination with different vaccines has been implemented globally to counter the continuous COVID-19 pandemic. However, the vaccine-elicited antibodies have reduced efficiency against the highly mutated Omicron sub-variants. Previously, we developed a protein subunit COVID-19 vaccine called ZF2001, based on the dimeric receptor-binding domain (RBD). This vaccine has been administered using different dosing intervals in real-world setting. Some individuals received three doses of ZF2001, with a one-month interval between each dose, due to urgent clinical requirements. Others had an extended dosing interval of up to five months between the second and third dose, a standard vaccination regimen for the protein subunit vaccine against hepatitis B. In this study, we profile B cell responses in individuals who received three doses of ZF2001, and compared those with long or short dosing intervals. We observed that the long-interval group exhibited higher and broader serologic antibody responses. These responses were associated with the increased size and evolution of vaccine-elicited B-cell receptor repertoires, characterized by the elevation of expanded clonotypes and somatic hypermutations. Both groups of individuals generated substantial amounts of broadly neutralizing antibodies (bnAbs) against various SARS-CoV-2 variants, including Omicron sub-variants such as XBB. These bnAbs target four antigenic sites within the RBD. To determine the vulnerable site of SARS-CoV-2, we employed cryo-electron microscopy to identify the epitopes of highly potent bnAbs that targeted two major sites. Our findings provide immunological insights into the B cell responses elicited by RBD-based vaccine, and suggest that a vaccination regimen of prolonging time interval should be used in practice. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_34944.map.gz emd_34944.map.gz | 483.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-34944-v30.xml emd-34944-v30.xml emd-34944.xml emd-34944.xml | 22 KB 22 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_34944.png emd_34944.png | 78.1 KB | ||
| Filedesc metadata |  emd-34944.cif.gz emd-34944.cif.gz | 6.6 KB | ||
| Others |  emd_34944_half_map_1.map.gz emd_34944_half_map_1.map.gz emd_34944_half_map_2.map.gz emd_34944_half_map_2.map.gz | 474.8 MB 474.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-34944 http://ftp.pdbj.org/pub/emdb/structures/EMD-34944 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34944 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34944 | HTTPS FTP |
-Validation report
| Summary document |  emd_34944_validation.pdf.gz emd_34944_validation.pdf.gz | 730.6 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_34944_full_validation.pdf.gz emd_34944_full_validation.pdf.gz | 730.2 KB | Display | |
| Data in XML |  emd_34944_validation.xml.gz emd_34944_validation.xml.gz | 18.1 KB | Display | |
| Data in CIF |  emd_34944_validation.cif.gz emd_34944_validation.cif.gz | 21.6 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34944 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34944 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34944 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34944 | HTTPS FTP |
-Related structure data
| Related structure data |  8hpuMC  8hp9C  8hpfC  8hpqC  8hpvC  8hq7C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_34944.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_34944.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-EM structure of SARS-CoV-2 Omicron BA.4 RBD in complex with fab L4.65 and L5.34 | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.84 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Cryo-EM structure of SARS-CoV-2 Omicron BA.4 RBD in...
| File | emd_34944_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-EM structure of SARS-CoV-2 Omicron BA.4 RBD in complex with fab L4.65 and L5.34 | ||||||||||||
| Projections & Slices |
| ||||||||||||
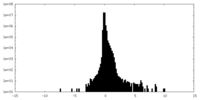
| Density Histograms |
-Half map: Cryo-EM structure of SARS-CoV-2 Omicron BA.4 RBD in...
| File | emd_34944_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-EM structure of SARS-CoV-2 Omicron BA.4 RBD in complex with fab L4.65 and L5.34 | ||||||||||||
| Projections & Slices |
| ||||||||||||
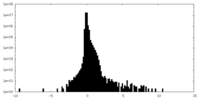
| Density Histograms |
- Sample components
Sample components
-Entire : Cryo-EM structure of SARS-CoV-2 Omicron BA.4 RBD in complex with ...
| Entire | Name: Cryo-EM structure of SARS-CoV-2 Omicron BA.4 RBD in complex with fab L4.65 and L5.34 |
|---|---|
| Components |
|
-Supramolecule #1: Cryo-EM structure of SARS-CoV-2 Omicron BA.4 RBD in complex with ...
| Supramolecule | Name: Cryo-EM structure of SARS-CoV-2 Omicron BA.4 RBD in complex with fab L4.65 and L5.34 type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#5 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Spike protein S2'
| Macromolecule | Name: Spike protein S2' / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 21.905697 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: NLCPFDEVFN ATRFASVYAW NRKRISNCVA DYSVLYNFAP FFAFKCYGVS PTKLNDLCFT NVYADSFVIR GNEVSQIAPG QTGNIADYN YKLPDDFTGC VIAWNSNKLD SKVGGNYNYR YRLFRKSNLK PFERDISTEI YQAGNKPCNG VAGVNCYFPL Q SYGFRPTY ...String: NLCPFDEVFN ATRFASVYAW NRKRISNCVA DYSVLYNFAP FFAFKCYGVS PTKLNDLCFT NVYADSFVIR GNEVSQIAPG QTGNIADYN YKLPDDFTGC VIAWNSNKLD SKVGGNYNYR YRLFRKSNLK PFERDISTEI YQAGNKPCNG VAGVNCYFPL Q SYGFRPTY GVGHQPYRVV VLSFELLHAP ATVCGP UniProtKB: Spike glycoprotein |
-Macromolecule #2: fab L4.65
| Macromolecule | Name: fab L4.65 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 24.953938 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QITLKESGPT LVKPTQTLTL TCTFSGFSLS TSGVGVAWIR QPPGKALEWL ALIYWDNDKR SSPSLNNRLT ITKDTSKNQV VLTMTNMDP EDTATYYCAH FFSHYDSSNY YYGSWFDPWG QGTLVTVSSA STKGPSVFPL APSSKSTSGG TAALGCLVKD Y FPEPVTVS ...String: QITLKESGPT LVKPTQTLTL TCTFSGFSLS TSGVGVAWIR QPPGKALEWL ALIYWDNDKR SSPSLNNRLT ITKDTSKNQV VLTMTNMDP EDTATYYCAH FFSHYDSSNY YYGSWFDPWG QGTLVTVSSA STKGPSVFPL APSSKSTSGG TAALGCLVKD Y FPEPVTVS WNSGALTSGV HTFPAVLQSS GLYSLSSVVT VPSSSLGTQT YICNVNHKPS NTKVDKRVEP KSC |
-Macromolecule #3: fab L4.65
| Macromolecule | Name: fab L4.65 / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 23.556061 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: EIVLTQSPGT LSLSPGERAT LSCRASQSFD SRYLGWYQQK SGQAPRLLIY GASSRATGIP DRFSGSGSGT DFTLTISRLE PEDFAVYYC QQFGDSPFTF GQGTKLEIKR TVAAPSVFIF PPSDEQLKSG TASVVCLLNN FYPREAKVQW KVDNALQSGN S QESVTEQD ...String: EIVLTQSPGT LSLSPGERAT LSCRASQSFD SRYLGWYQQK SGQAPRLLIY GASSRATGIP DRFSGSGSGT DFTLTISRLE PEDFAVYYC QQFGDSPFTF GQGTKLEIKR TVAAPSVFIF PPSDEQLKSG TASVVCLLNN FYPREAKVQW KVDNALQSGN S QESVTEQD SKDSTYSLSS TLTLSKADYE KHKVYACEVT HQGLSSPVTK SFNRGEC |
-Macromolecule #4: fab L5.34
| Macromolecule | Name: fab L5.34 / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 23.592424 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: VQLVESGGGL VQPGGSLRLS CAASEITVSS NYMNWVRQAP GKGLEWVSVV YPGGSTFYTD SVKGRFTISR DNSKNTLYLQ MNSLRAEDT AVYYCARESG GFPLAEGAFD IWGQGTMVTV SSASTKGPSV FPLAPSSKST SGGTAALGCL VKDYFPEPVT V SWNSGALT ...String: VQLVESGGGL VQPGGSLRLS CAASEITVSS NYMNWVRQAP GKGLEWVSVV YPGGSTFYTD SVKGRFTISR DNSKNTLYLQ MNSLRAEDT AVYYCARESG GFPLAEGAFD IWGQGTMVTV SSASTKGPSV FPLAPSSKST SGGTAALGCL VKDYFPEPVT V SWNSGALT SGVHTFPAVL QSSGLYSLSS VVTVPSSSLG TQTYICNVNH KPSNTKVDKR VEPKSC |
-Macromolecule #5: fab L5.34
| Macromolecule | Name: fab L5.34 / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 23.379928 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: DIQMTQSPSS LSASVGDRVS ITCRASQSIS THLHWYQQKP GKAPKLLISA ASTLQSGVPS RFSGSGSGTD FTLTITSLQP EDFATYYCQ QSYSTPRGLS FGGGTKVEIK RTVAAPSVFI FPPSDEQLKS GTASVVCLLN NFYPREAKVQ WKVDNALQSG N SQESVTEQ ...String: DIQMTQSPSS LSASVGDRVS ITCRASQSIS THLHWYQQKP GKAPKLLISA ASTLQSGVPS RFSGSGSGTD FTLTITSLQP EDFATYYCQ QSYSTPRGLS FGGGTKVEIK RTVAAPSVFI FPPSDEQLKS GTASVVCLLN NFYPREAKVQ WKVDNALQSG N SQESVTEQ DSKDSTYSLS STLTLSKADY EKHKVYACEV THQGLSSPVT KSFNRGEC |
-Macromolecule #6: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 6 / Number of copies: 1 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Average electron dose: 1.39 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 5.0 µm / Nominal defocus min: 1.2 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)