[English] 日本語
 Yorodumi
Yorodumi- EMDB-33538: Cryo-EM structure of human IgM-Fc in complex with the J chain and... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of human IgM-Fc in complex with the J chain and the DBL domain of DBLMSP | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | malaria / immunoglobin / IMMUNE SYSTEM | |||||||||
| Function / homology |  Function and homology information Function and homology informationhexameric IgM immunoglobulin complex / dimeric IgA immunoglobulin complex / IgM B cell receptor complex / secretory dimeric IgA immunoglobulin complex / pentameric IgM immunoglobulin complex / monomeric IgA immunoglobulin complex / secretory IgA immunoglobulin complex / IgA binding / pre-B cell allelic exclusion / IgM immunoglobulin complex ...hexameric IgM immunoglobulin complex / dimeric IgA immunoglobulin complex / IgM B cell receptor complex / secretory dimeric IgA immunoglobulin complex / pentameric IgM immunoglobulin complex / monomeric IgA immunoglobulin complex / secretory IgA immunoglobulin complex / IgA binding / pre-B cell allelic exclusion / IgM immunoglobulin complex / glomerular filtration / CD22 mediated BCR regulation / immunoglobulin complex, circulating / immunoglobulin receptor binding / positive regulation of respiratory burst / humoral immune response / Scavenging of heme from plasma / complement activation, classical pathway / antigen binding / Antigen activates B Cell Receptor (BCR) leading to generation of second messengers / Cell surface interactions at the vascular wall / B cell receptor signaling pathway / antibacterial humoral response / protein-macromolecule adaptor activity / protein-containing complex assembly / defense response to Gram-negative bacterium / blood microparticle / adaptive immune response / Potential therapeutics for SARS / host cell surface receptor binding / immune response / innate immune response / cell surface / protein homodimerization activity / extracellular space / extracellular exosome / extracellular region / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.71 Å | |||||||||
 Authors Authors | Shen H / Ji C / Xiao J | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: Plasmodium falciparum has evolved multiple mechanisms to hijack human immunoglobulin M. Authors: Chenggong Ji / Hao Shen / Chen Su / Yaxin Li / Shihua Chen / Thomas H Sharp / Junyu Xiao /   Abstract: Plasmodium falciparum causes the most severe malaria in humans. Immunoglobulin M (IgM) serves as the first line of humoral defense against infection and potently activates the complement pathway to ...Plasmodium falciparum causes the most severe malaria in humans. Immunoglobulin M (IgM) serves as the first line of humoral defense against infection and potently activates the complement pathway to facilitate P. falciparum clearance. A number of P. falciparum proteins bind IgM, leading to immune evasion and severe disease. However, the underlying molecular mechanisms remain unknown. Here, using high-resolution cryo-electron microscopy, we delineate how P. falciparum proteins VAR2CSA, TM284VAR1, DBLMSP, and DBLMSP2 target IgM. Each protein binds IgM in a different manner, and together they present a variety of Duffy-binding-like domain-IgM interaction modes. We further show that these proteins interfere directly with IgM-mediated complement activation in vitro, with VAR2CSA exhibiting the most potent inhibitory effect. These results underscore the importance of IgM for human adaptation of P. falciparum and provide critical insights into its immune evasion mechanism. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_33538.map.gz emd_33538.map.gz | 118.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-33538-v30.xml emd-33538-v30.xml emd-33538.xml emd-33538.xml | 18 KB 18 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_33538.png emd_33538.png | 63.2 KB | ||
| Filedesc metadata |  emd-33538.cif.gz emd-33538.cif.gz | 6.2 KB | ||
| Others |  emd_33538_half_map_1.map.gz emd_33538_half_map_1.map.gz emd_33538_half_map_2.map.gz emd_33538_half_map_2.map.gz | 115.8 MB 115.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-33538 http://ftp.pdbj.org/pub/emdb/structures/EMD-33538 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33538 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33538 | HTTPS FTP |
-Validation report
| Summary document |  emd_33538_validation.pdf.gz emd_33538_validation.pdf.gz | 1.1 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_33538_full_validation.pdf.gz emd_33538_full_validation.pdf.gz | 1.1 MB | Display | |
| Data in XML |  emd_33538_validation.xml.gz emd_33538_validation.xml.gz | 13.9 KB | Display | |
| Data in CIF |  emd_33538_validation.cif.gz emd_33538_validation.cif.gz | 16.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33538 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33538 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33538 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33538 | HTTPS FTP |
-Related structure data
| Related structure data |  7y09MC  7y0hC  7y0jC  7yg2C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_33538.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_33538.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.08 Å | ||||||||||||||||||||||||||||||||||||
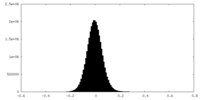
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_33538_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
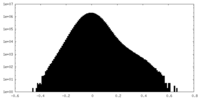
| Density Histograms |
-Half map: #1
| File | emd_33538_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
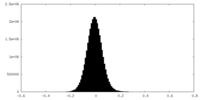
| Density Histograms |
- Sample components
Sample components
-Entire : Ternary complex of human IgM-Fc with the J chain and the DBL doma...
| Entire | Name: Ternary complex of human IgM-Fc with the J chain and the DBL domain of DBLMSP |
|---|---|
| Components |
|
-Supramolecule #1: Ternary complex of human IgM-Fc with the J chain and the DBL doma...
| Supramolecule | Name: Ternary complex of human IgM-Fc with the J chain and the DBL domain of DBLMSP type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Source (natural) | Organism:  |
-Supramolecule #2: Putative erythrocyte membrane protein
| Supramolecule | Name: Putative erythrocyte membrane protein / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Supramolecule #3: Immunoglobulin heavy constant mu
| Supramolecule | Name: Immunoglobulin heavy constant mu / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #2 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Supramolecule #4: Immunoglobulin J chain
| Supramolecule | Name: Immunoglobulin J chain / type: complex / ID: 4 / Parent: 1 / Macromolecule list: #3 |
|---|
-Macromolecule #1: Putative erythrocyte membrane protein
| Macromolecule | Name: Putative erythrocyte membrane protein / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 75.107406 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: DSNLRNGLLN NSLDLTNGLN NKDNSFIDSK IEEHENKSYQ NKDNNISIVG QDVPITSVDS SKIINANDLE GNSIDDTKGL SVTNSGFDD GSAFGGGLPF SGYSPLQGNH NKCPDENFCK GIKNVLSCPP KNSTGRNGDW ISVAVKESST TNKGVLVPPR R KKLCLRNI ...String: DSNLRNGLLN NSLDLTNGLN NKDNSFIDSK IEEHENKSYQ NKDNNISIVG QDVPITSVDS SKIINANDLE GNSIDDTKGL SVTNSGFDD GSAFGGGLPF SGYSPLQGNH NKCPDENFCK GIKNVLSCPP KNSTGRNGDW ISVAVKESST TNKGVLVPPR R KKLCLRNI NQVWHRIKDE KNFKEEFVKV ALGESNALMK HYKEKNLNAL TAIKYGFSDM GDIIKGTDLI DYQITKNINR AL DKILGNE TSNDKIKKRV DWWEANKSAF WDAFMCGYKV HIGNKPCPEH DNMDRIPQYL RWFREWGTYV CSEYKNKFEN VIE LCNVRQ ITNQNDSQLL EISKEDKCKG ALKHYEEWVN RRRPEWKGQC DKFEKEKSKY EDTKSRTAEI YLKEICSECD CKYK DLDNT FKEFKDNVTL LKAVIDNKKN QDSLTTTSLS TSINSVRDSS NLDQRGNITT SQGNSHRATV VQQADQTNRL DNVNS VTQR GNNNYNNNLE RGLGSGALPG TNIITEEKYS LELIKLTSKD EEDIIKHNED VREEIEEQQE DIEEDEEELE NEGEET KEE DDEEKNETND TEDTDDTEDT EDIEEENEEK ELSNQQQSEK KSISKVDEDS YRILSVSYKD NNEVKNVAES IVKKLFS LF NDNNNLETIF KGLT UniProtKB: Putative erythrocyte membrane protein |
-Macromolecule #2: Immunoglobulin heavy constant mu
| Macromolecule | Name: Immunoglobulin heavy constant mu / type: protein_or_peptide / ID: 2 / Number of copies: 10 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 41.875766 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: ASAWSHPQFE KGGGSGGGSG GSAWSHPQFE KIDTTIAELP PKVSVFVPPR DGFFGNPRKS KLICQATGFS PRQIQVSWLR EGKQVGSGV TTDQVQAEAK ESGPTTYKVT STLTIKESDW LGQSMFTCRV DHRGLTFQQN ASSMCVPDQD TAIRVFAIPP S FASIFLTK ...String: ASAWSHPQFE KGGGSGGGSG GSAWSHPQFE KIDTTIAELP PKVSVFVPPR DGFFGNPRKS KLICQATGFS PRQIQVSWLR EGKQVGSGV TTDQVQAEAK ESGPTTYKVT STLTIKESDW LGQSMFTCRV DHRGLTFQQN ASSMCVPDQD TAIRVFAIPP S FASIFLTK STKLTCLVTD LTTYDSVTIS WTRQNGEAVK THTNISESHP NATFSAVGEA SICEDDWNSG ERFTCTVTHT DL PSPLKQT ISRPKGVALH RPDVYLLPPA REQLNLRESA TITCLVTGFS PADVFVQWMQ RGQPLSPEKY VTSAPMPEPQ APG RYFAHS ILTVSEEEWN TGETYTCVVA HEALPNRVTE RTVDKSTGKP TLYNVSLVMS DTAGTCY UniProtKB: Immunoglobulin heavy constant mu |
-Macromolecule #3: Immunoglobulin J chain
| Macromolecule | Name: Immunoglobulin J chain / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 15.483329 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: EDERIVLVDN KCKCARITSR IIRSSEDPNE DIVERNIRII VPLNNRENIS DPTSPLRTRF VYHLSDLCKK CDPTEVELDN QIVTATQSN ICDEDSATET CYTYDRNKCY TAVVPLVYGG ETKMVETALT PDACYPD UniProtKB: Immunoglobulin J chain |
-Macromolecule #5: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 5 / Number of copies: 10 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.5 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: PDB ENTRY PDB model - PDB ID: |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.71 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 391618 |
| Initial angle assignment | Type: ANGULAR RECONSTITUTION |
| Final angle assignment | Type: ANGULAR RECONSTITUTION |
 Movie
Movie Controller
Controller























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)