+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM map of E.coli FtsH AAA protease | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 4.2 Å | |||||||||
 Authors Authors | Qiao Z / Gao YG | |||||||||
| Funding support |  Singapore, 1 items Singapore, 1 items
| |||||||||
 Citation Citation |  Journal: Cell Rep / Year: 2022 Journal: Cell Rep / Year: 2022Title: Cryo-EM structure of the entire FtsH-HflKC AAA protease complex. Authors: Zhu Qiao / Tatsuhiko Yokoyama / Xin-Fu Yan / Ing Tsyr Beh / Jian Shi / Sandip Basak / Yoshinori Akiyama / Yong-Gui Gao /   Abstract: The membrane-bound AAA protease FtsH is the key player controlling protein quality in bacteria. Two single-pass membrane proteins, HflK and HflC, interact with FtsH to modulate its proteolytic ...The membrane-bound AAA protease FtsH is the key player controlling protein quality in bacteria. Two single-pass membrane proteins, HflK and HflC, interact with FtsH to modulate its proteolytic activity. Here, we present structure of the entire FtsH-HflKC complex, comprising 12 copies of both HflK and HflC, all of which interact reciprocally to form a cage, as well as four FtsH hexamers with periplasmic domains and transmembrane helices enclosed inside the cage and cytoplasmic domains situated at the base of the cage. FtsH K61/D62/S63 in the β2-β3 loop in the periplasmic domain directly interact with HflK, contributing to complex formation. Pull-down and in vivo enzymatic activity assays validate the importance of the interacting interface for FtsH-HflKC complex formation. Structural comparison with the substrate-bound human m-AAA protease AFG3L2 offers implications for the HflKC cage in modulating substrate access to FtsH. Together, our findings provide a better understanding of FtsH-type AAA protease holoenzyme assembly and regulation. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_32524.map.gz emd_32524.map.gz | 11.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-32524-v30.xml emd-32524-v30.xml emd-32524.xml emd-32524.xml | 9 KB 9 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_32524.png emd_32524.png | 36.6 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-32524 http://ftp.pdbj.org/pub/emdb/structures/EMD-32524 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-32524 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-32524 | HTTPS FTP |
-Validation report
| Summary document |  emd_32524_validation.pdf.gz emd_32524_validation.pdf.gz | 390.7 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_32524_full_validation.pdf.gz emd_32524_full_validation.pdf.gz | 390.2 KB | Display | |
| Data in XML |  emd_32524_validation.xml.gz emd_32524_validation.xml.gz | 5.5 KB | Display | |
| Data in CIF |  emd_32524_validation.cif.gz emd_32524_validation.cif.gz | 6.2 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-32524 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-32524 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-32524 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-32524 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_32524.map.gz / Format: CCP4 / Size: 15.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_32524.map.gz / Format: CCP4 / Size: 15.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.3728 Å | ||||||||||||||||||||||||||||||||||||
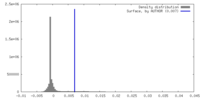
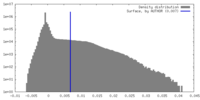
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : FtsH
| Entire | Name: FtsH |
|---|---|
| Components |
|
-Supramolecule #1: FtsH
| Supramolecule | Name: FtsH / type: complex / Chimera: Yes / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
-Macromolecule #1: E.Coli FtsH AAA protease
| Macromolecule | Name: E.Coli FtsH AAA protease / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
| Sequence | String: MAKNLILWLV IAVVLMSVFQ SFGPSESNGR KVDYSTFLQE VNNDQVREAR INGREINVTK KDSNRYTTY IPVQDPKLLD NLLTKNVKVV GEPPEEPSLL ASIFISWFPM LLLIGVWIFF M RQMQGGGG KGAMSFGKSK ARMLTEDQIK TTFADVAGCD EAKEEVAELV ...String: MAKNLILWLV IAVVLMSVFQ SFGPSESNGR KVDYSTFLQE VNNDQVREAR INGREINVTK KDSNRYTTY IPVQDPKLLD NLLTKNVKVV GEPPEEPSLL ASIFISWFPM LLLIGVWIFF M RQMQGGGG KGAMSFGKSK ARMLTEDQIK TTFADVAGCD EAKEEVAELV EYLREPSRFQ KL GGKIPKG VLMVGPPGTG KTLLAKAIAG EAKVPFFTIS GSDFVEMFVG VGASRVRDMF EQA KKAAPC IIFIDEIDAV GRQRGAGLGG GHDEREQTLN QMLVEMDGFE GNEGIIVIAA TNRP DVLDP ALLRPGRFDR QVVVGLPDVR GREQILKVHM RRVPLAPDID AAIIARGTPG FSGAD LANL VNEAALFAAR GNKRVVSMVE FEKAKDKIMM GAERRSMVMT EAQKESTAYH EAGHAI IGR LVPEHDPVHK VTIIPRGRAL GVTFFLPEGD AISASRQKLE SQISTLYGGR LAEEIIY GP EHVSTGASND IKVATNLARN MVTQWGFSEK LGPLLYAEEE GEVFLGRSVA KAKHMSDE T ARIIDQEVKA LIERNYNRAR QLLTDNMDIL HAMKDALMKY ETIDAPQIDD LMARRDVRP PAGWEEPGAS NNSGDNGSPK APRPVDEPRT PNPGNTMSEQ LGDK |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.6 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.5 µm / Nominal defocus min: 0.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 4.2 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 40539 |
|---|---|
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)