[English] 日本語
 Yorodumi
Yorodumi- EMDB-29648: Cryo-EM structure of SQ31f-bound Mycobacterium smegmatis ATP synt... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of SQ31f-bound Mycobacterium smegmatis ATP synthase FO region | |||||||||
 Map data Map data | Cryo-EM structure of SQ31f-bound Mycobacterium smegmatis ATP synthase FO region (sharpened) | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | ATP synthase / mycobacterial / inhibitor / tuberculosis / antibiotic / HYDROLASE / TRANSLOCASE-INHIBITOR complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationproton-transporting ATP synthase complex, coupling factor F(o) / proton-transporting ATP synthase activity, rotational mechanism / hydrolase activity / lipid binding / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Mycolicibacterium smegmatis MC2 155 (bacteria) Mycolicibacterium smegmatis MC2 155 (bacteria) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.8 Å | |||||||||
 Authors Authors | Courbon GM / Rubinstein JL | |||||||||
| Funding support |  Canada, 1 items Canada, 1 items
| |||||||||
 Citation Citation |  Journal: EMBO J / Year: 2023 Journal: EMBO J / Year: 2023Title: Mechanism of mycobacterial ATP synthase inhibition by squaramides and second generation diarylquinolines. Authors: Gautier M Courbon / Paul R Palme / Lea Mann / Adrian Richter / Peter Imming / John L Rubinstein /   Abstract: Mycobacteria, such as Mycobacterium tuberculosis, depend on the activity of adenosine triphosphate (ATP) synthase for growth. The diarylquinoline bedaquiline (BDQ), a mycobacterial ATP synthase ...Mycobacteria, such as Mycobacterium tuberculosis, depend on the activity of adenosine triphosphate (ATP) synthase for growth. The diarylquinoline bedaquiline (BDQ), a mycobacterial ATP synthase inhibitor, is an important medication for treatment of drug-resistant tuberculosis but suffers from off-target effects and is susceptible to resistance mutations. Consequently, both new and improved mycobacterial ATP synthase inhibitors are needed. We used electron cryomicroscopy and biochemical assays to study the interaction of Mycobacterium smegmatis ATP synthase with the second generation diarylquinoline TBAJ-876 and the squaramide inhibitor SQ31f. The aryl groups of TBAJ-876 improve binding compared with BDQ, while SQ31f, which blocks ATP synthesis ~10 times more potently than ATP hydrolysis, binds a previously unknown site in the enzyme's proton-conducting channel. Remarkably, BDQ, TBAJ-876, and SQ31f all induce similar conformational changes in ATP synthase, suggesting that the resulting conformation is particularly suited for drug binding. Further, high concentrations of the diarylquinolines uncouple the transmembrane proton motive force while for SQ31f they do not, which may explain why high concentrations of diarylquinolines, but not SQ31f, have been reported to kill mycobacteria. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_29648.map.gz emd_29648.map.gz | 118 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-29648-v30.xml emd-29648-v30.xml emd-29648.xml emd-29648.xml | 18.7 KB 18.7 KB | Display Display |  EMDB header EMDB header |
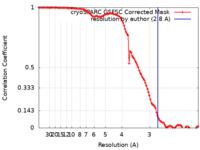
| FSC (resolution estimation) |  emd_29648_fsc.xml emd_29648_fsc.xml | 10.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_29648.png emd_29648.png | 91.9 KB | ||
| Filedesc metadata |  emd-29648.cif.gz emd-29648.cif.gz | 5.9 KB | ||
| Others |  emd_29648_half_map_1.map.gz emd_29648_half_map_1.map.gz emd_29648_half_map_2.map.gz emd_29648_half_map_2.map.gz | 115.9 MB 115.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-29648 http://ftp.pdbj.org/pub/emdb/structures/EMD-29648 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29648 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29648 | HTTPS FTP |
-Validation report
| Summary document |  emd_29648_validation.pdf.gz emd_29648_validation.pdf.gz | 871.8 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_29648_full_validation.pdf.gz emd_29648_full_validation.pdf.gz | 871.4 KB | Display | |
| Data in XML |  emd_29648_validation.xml.gz emd_29648_validation.xml.gz | 18.9 KB | Display | |
| Data in CIF |  emd_29648_validation.cif.gz emd_29648_validation.cif.gz | 24.1 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-29648 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-29648 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-29648 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-29648 | HTTPS FTP |
-Related structure data
| Related structure data |  8g07MC  8g08C  8g09C  8g0aC  8g0bC  8g0cC  8g0dC  8g0eC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_29648.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_29648.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-EM structure of SQ31f-bound Mycobacterium smegmatis ATP synthase FO region (sharpened) | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.03 Å | ||||||||||||||||||||||||||||||||||||
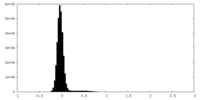
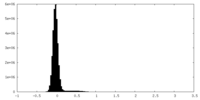
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Cryo-EM structure of SQ31f-bound Mycobacterium smegmatis ATP synthase...
| File | emd_29648_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-EM structure of SQ31f-bound Mycobacterium smegmatis ATP synthase FO region (Half A) | ||||||||||||
| Projections & Slices |
| ||||||||||||
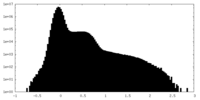
| Density Histograms |
-Half map: Cryo-EM structure of SQ31f-bound Mycobacterium smegmatis ATP synthase...
| File | emd_29648_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-EM structure of SQ31f-bound Mycobacterium smegmatis ATP synthase FO region (Half B) | ||||||||||||
| Projections & Slices |
| ||||||||||||
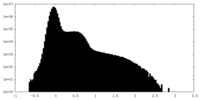
| Density Histograms |
- Sample components
Sample components
-Entire : SQ31f-bound Mycobacterium smegmatis ATP synthase FO region
| Entire | Name: SQ31f-bound Mycobacterium smegmatis ATP synthase FO region |
|---|---|
| Components |
|
-Supramolecule #1: SQ31f-bound Mycobacterium smegmatis ATP synthase FO region
| Supramolecule | Name: SQ31f-bound Mycobacterium smegmatis ATP synthase FO region type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 |
|---|---|
| Source (natural) | Organism:  Mycolicibacterium smegmatis MC2 155 (bacteria) Mycolicibacterium smegmatis MC2 155 (bacteria) |
-Macromolecule #1: ATP synthase subunit c
| Macromolecule | Name: ATP synthase subunit c / type: protein_or_peptide / ID: 1 / Number of copies: 9 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Mycolicibacterium smegmatis MC2 155 (bacteria) Mycolicibacterium smegmatis MC2 155 (bacteria) |
| Molecular weight | Theoretical: 8.597982 KDa |
| Sequence | String: MDLDPNAIIT AGALIGGGLI MGGGAIGAGI GDGIAGNALI SGIARQPEAQ GRLFTPFFIT VGLVEAAYFI NLAFMALFVF ATPGLQ UniProtKB: ATP synthase subunit c |
-Macromolecule #2: ATP synthase subunit a
| Macromolecule | Name: ATP synthase subunit a / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Mycolicibacterium smegmatis MC2 155 (bacteria) Mycolicibacterium smegmatis MC2 155 (bacteria) |
| Molecular weight | Theoretical: 27.568482 KDa |
| Sequence | String: MLAAEEGGAA IHVGHHTLVF ELFGMTFNGD TILATAVTAV IVIALAFYLR AKVTSTGVPS GVQLFWEALT IQMRQQIEGS IGMKIAPFV LPLSVTIFVF ILISNWLAVL PLQYGGADGA AAELYKAPAS DINFVLALAL FVFVCYHAAG IWRRGIVGHP I KVVKGHVA ...String: MLAAEEGGAA IHVGHHTLVF ELFGMTFNGD TILATAVTAV IVIALAFYLR AKVTSTGVPS GVQLFWEALT IQMRQQIEGS IGMKIAPFV LPLSVTIFVF ILISNWLAVL PLQYGGADGA AAELYKAPAS DINFVLALAL FVFVCYHAAG IWRRGIVGHP I KVVKGHVA FLAPINIVEE LAKPISLALR LFGNIFAGGI LVALIAMFPW YIQWFPNAVW KTFDLFVGLI QAFIFSLLTI LY FSQSMEL DHEDH UniProtKB: ATP synthase subunit a |
-Macromolecule #3: ATP synthase subunit b
| Macromolecule | Name: ATP synthase subunit b / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Mycolicibacterium smegmatis MC2 155 (bacteria) Mycolicibacterium smegmatis MC2 155 (bacteria) |
| Molecular weight | Theoretical: 17.636701 KDa |
| Sequence | String: MGEFSATILA ASQAAEEGGG GSNFLIPNGT FFAVLIIFLI VLGVISKWVV PPISKVLAER EAMLAKTAAD NRKSAEQVAA AQADYEKEM AEARAQASAL RDEARAAGRS VVDEKRAQAS GEVAQTLTQA DQQLSAQGDQ VRSGLESSVD GLSAKLASRI L GVDVNSGG TQ UniProtKB: ATP synthase subunit b |
-Macromolecule #4: ATP synthase subunit b-delta
| Macromolecule | Name: ATP synthase subunit b-delta / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Mycolicibacterium smegmatis MC2 155 (bacteria) Mycolicibacterium smegmatis MC2 155 (bacteria) |
| Molecular weight | Theoretical: 47.504723 KDa |
| Sequence | String: MSIFIGQLIG FAVIAFIIVK WVVPPVRTLM RNQQEAVRAA LAESAEAAKK LADADAMHAK ALADAKAESE KVTEEAKQDS ERIAAQLSE QAGSEAERIK AQGAQQIQLM RQQLIRQLRT GLGAEAVNKA AEIVRAHVAD PQAQSATVDR FLSELEQMAP S SVVIDTAA ...String: MSIFIGQLIG FAVIAFIIVK WVVPPVRTLM RNQQEAVRAA LAESAEAAKK LADADAMHAK ALADAKAESE KVTEEAKQDS ERIAAQLSE QAGSEAERIK AQGAQQIQLM RQQLIRQLRT GLGAEAVNKA AEIVRAHVAD PQAQSATVDR FLSELEQMAP S SVVIDTAA TSRLRAASRQ SLAALVEKFD SVAGGLDADG LTNLADELAS VAKLLLSETA LNKHLAEPTD DSAPKVRLLE RL LSDKVSA TTLDLLRTAV SNRWSTESNL IDAVEHTARL ALLKRAEIAG EVDEVEEQLF RFGRVLDAEP RLSALLSDYT TPA EGRVAL LDKALTGRPG VNQTAAALLS QTVGLLRGER ADEAVIDLAE LAVSRRGEVV AHVSAAAELS DAQRTRLTEV LSRI YGRPV SVQLHVDPEL LGGLSITVGD EVIDGSIASR LAAAQTGLPD UniProtKB: ATP synthase subunit b-delta |
-Macromolecule #5: 3-[4-(morpholin-4-yl)phenyl]-4-{[(pyridin-2-yl)methyl]amino}cyclo...
| Macromolecule | Name: 3-[4-(morpholin-4-yl)phenyl]-4-{[(pyridin-2-yl)methyl]amino}cyclobut-3-ene-1,2-dione type: ligand / ID: 5 / Number of copies: 1 / Formula: SQC |
|---|---|
| Molecular weight | Theoretical: 349.383 Da |
| Chemical component information |  ChemComp-SQC: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Average electron dose: 54.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.9 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)