+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | YES Complex - E. coli MraY, Protein E ID21, E. coli SlyD | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | inhibitor / antibiotic / chaperone / membrane / bacteriophage / TRANSFERASE-ISOMERASE complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationsuppression by virus of host peptidoglycan biosynthetic process / viral release via suppression of host peptidoglycan biosynthetic process / phospho-N-acetylmuramoyl-pentapeptide-transferase / UDP-N-acetylmuramoyl-L-alanyl-D-glutamyl-meso-2,6-diaminopimelyl-D-alanyl-D-alanine:undecaprenyl-phosphate transferase activity / phospho-N-acetylmuramoyl-pentapeptide-transferase activity / cell wall macromolecule biosynthetic process / enzyme inhibitor activity / peptidoglycan biosynthetic process / viral release from host cell by cytolysis / peptidylprolyl isomerase ...suppression by virus of host peptidoglycan biosynthetic process / viral release via suppression of host peptidoglycan biosynthetic process / phospho-N-acetylmuramoyl-pentapeptide-transferase / UDP-N-acetylmuramoyl-L-alanyl-D-glutamyl-meso-2,6-diaminopimelyl-D-alanyl-D-alanine:undecaprenyl-phosphate transferase activity / phospho-N-acetylmuramoyl-pentapeptide-transferase activity / cell wall macromolecule biosynthetic process / enzyme inhibitor activity / peptidoglycan biosynthetic process / viral release from host cell by cytolysis / peptidylprolyl isomerase / peptidyl-prolyl cis-trans isomerase activity / cell wall organization / protein folding / regulation of cell shape / killing of cells of another organism / cell cycle / cell division / host cell plasma membrane / membrane / metal ion binding / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Escherichia phage phiX174 (virus) / Escherichia phage phiX174 (virus) /  | |||||||||
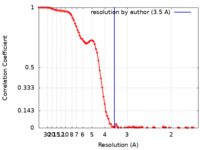
| Method | single particle reconstruction / cryo EM / Resolution: 3.5 Å | |||||||||
 Authors Authors | Orta AK / Clemons WM / Li YE | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Science / Year: 2023 Journal: Science / Year: 2023Title: The mechanism of the phage-encoded protein antibiotic from ΦX174. Authors: Anna K Orta / Nadia Riera / Yancheng E Li / Shiho Tanaka / Hyun Gi Yun / Lada Klaic / William M Clemons /  Abstract: The historically important phage ΦX174 kills its host bacteria by encoding a 91-residue protein antibiotic called protein E. Using single-particle electron cryo-microscopy, we demonstrate that ...The historically important phage ΦX174 kills its host bacteria by encoding a 91-residue protein antibiotic called protein E. Using single-particle electron cryo-microscopy, we demonstrate that protein E bridges two bacterial proteins to form the transmembrane YES complex [MraY, protein E, sensitivity to lysis D (SlyD)]. Protein E inhibits peptidoglycan biosynthesis by obstructing the MraY active site leading to loss of lipid I production. We experimentally validate this result for two different viral species, providing a clear model for bacterial lysis and unifying previous experimental data. Additionally, we characterize the MraY structure-revealing features of this essential enzyme-and the structure of the chaperone SlyD bound to a protein. Our structures provide insights into the mechanism of phage-mediated lysis and for structure-based design of phage therapeutics. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_29642.map.gz emd_29642.map.gz | 55.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-29642-v30.xml emd-29642-v30.xml emd-29642.xml emd-29642.xml | 23.4 KB 23.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_29642_fsc.xml emd_29642_fsc.xml | 11.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_29642.png emd_29642.png | 95.5 KB | ||
| Filedesc metadata |  emd-29642.cif.gz emd-29642.cif.gz | 6.7 KB | ||
| Others |  emd_29642_half_map_1.map.gz emd_29642_half_map_1.map.gz emd_29642_half_map_2.map.gz emd_29642_half_map_2.map.gz | 59.1 MB 59.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-29642 http://ftp.pdbj.org/pub/emdb/structures/EMD-29642 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29642 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29642 | HTTPS FTP |
-Validation report
| Summary document |  emd_29642_validation.pdf.gz emd_29642_validation.pdf.gz | 808.2 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_29642_full_validation.pdf.gz emd_29642_full_validation.pdf.gz | 807.8 KB | Display | |
| Data in XML |  emd_29642_validation.xml.gz emd_29642_validation.xml.gz | 16 KB | Display | |
| Data in CIF |  emd_29642_validation.cif.gz emd_29642_validation.cif.gz | 21 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-29642 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-29642 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-29642 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-29642 | HTTPS FTP |
-Related structure data
| Related structure data |  8g02MC  8g01C C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_29642.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_29642.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Voxel size | X=Y=Z: 0.832 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_29642_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: CryoEM map of the YES complex (phix174)
| File | emd_29642_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | CryoEM map of the YES complex (phix174) | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : YES complex
| Entire | Name: YES complex |
|---|---|
| Components |
|
-Supramolecule #1: YES complex
| Supramolecule | Name: YES complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all Details: Hexameric complex of E. coli MraY dimer bound to two molecules of Protein E (phix174), stabilized by E. coli SlyD |
|---|---|
| Molecular weight | Theoretical: 140.32 KDa |
-Supramolecule #2: Lysis Protein E
| Supramolecule | Name: Lysis Protein E / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #2 / Details: Protein E from PhiX174 phage |
|---|---|
| Source (natural) | Organism:  Escherichia phage phiX174 (virus) Escherichia phage phiX174 (virus) |
| Molecular weight | Theoretical: 11.52476 KDa |
-Supramolecule #3: Dimeric structure of MraY
| Supramolecule | Name: Dimeric structure of MraY / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 39.88 KDa |
-Supramolecule #4: SlyD
| Supramolecule | Name: SlyD / type: complex / ID: 4 / Parent: 1 / Macromolecule list: #3 / Details: E. coli SlyD truncated at residue 154 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 20.86 KDa |
-Macromolecule #1: Phospho-N-acetylmuramoyl-pentapeptide-transferase
| Macromolecule | Name: Phospho-N-acetylmuramoyl-pentapeptide-transferase / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO EC number: phospho-N-acetylmuramoyl-pentapeptide-transferase |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 39.909539 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MLVWLAEHLV KYYSGFNVFS YLTFRAIVSL LTALFISLWM GPRMIAHLQK LSFGQVVRND GPESHFSKRG TPTMGGIMIL TAIVISVLL WAYPSNPYVW CVLVVLVGYG VIGFVDDYRK VVRKDTKGLI ARWKYFWMSV IALGVAFALY LAGKDTPATQ L VVPFFKDV ...String: MLVWLAEHLV KYYSGFNVFS YLTFRAIVSL LTALFISLWM GPRMIAHLQK LSFGQVVRND GPESHFSKRG TPTMGGIMIL TAIVISVLL WAYPSNPYVW CVLVVLVGYG VIGFVDDYRK VVRKDTKGLI ARWKYFWMSV IALGVAFALY LAGKDTPATQ L VVPFFKDV MPQLGLFYIL LAYFVIVGTG NAVNLTDGLD GLAIMPTVFV AGGFALVAWA TGNMNFASYL HIPYLRHAGE LV IVCTAIV GAGLGFLWFN TYPAQVFMGD VGSLALGGAL GIIAVLLRQE FLLVIMGGVF VVETLSVILQ VGSFKLRGQR IFR MAPIHH HYELKGWPEP RVIVRFWIIS LMLVLIGLAT LKVR UniProtKB: Phospho-N-acetylmuramoyl-pentapeptide-transferase |
-Macromolecule #2: Lysis protein E
| Macromolecule | Name: Lysis protein E / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Escherichia phage phiX174 (virus) Escherichia phage phiX174 (virus) |
| Molecular weight | Theoretical: 11.545908 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGVRWTLWDT LAFLLLLSLL LPSLLIMFIP STFKRPVSSW KARNLRKTLL MASSVRLKPL NCSRLPCVYA QETLTFLLTQ KKTCVKNYV RKEHHHHHH UniProtKB: Lysis protein E |
-Macromolecule #3: Peptidyl-prolyl cis-trans isomerase
| Macromolecule | Name: Peptidyl-prolyl cis-trans isomerase / type: protein_or_peptide / ID: 3 / Number of copies: 2 / Enantiomer: LEVO / EC number: peptidylprolyl isomerase |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 16.659486 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MKVAKDLVVS LAYQVRTEDG VLVDESPVSA PLDYLHGHGS LISGLETALE GHEVGDKFDV AVGANDAYGQ YDENLVQRVP KDVFMGVDE LQVGMRFLAE TDQGPVPVEI TAVEDDHVVV DGNHMLAGQN LKFNVEVVAI REATEEELAH GHVHG UniProtKB: Peptidyl-prolyl cis-trans isomerase |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 5 mg/mL | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
Details: Supplemented with 2mM E. coli lipid extract | ||||||||||||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY | ||||||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277.15 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number grids imaged: 1 / Number real images: 10798 / Average exposure time: 2.0 sec. / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD / Nominal defocus max: 5.0 µm / Nominal defocus min: 0.5 µm / Nominal magnification: 105000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller







 Z
Z Y
Y X
X