+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | KWOCA 4 nanoparticle | |||||||||
 Map data Map data | KWOCA 4 nanoparticle - The main cryoEM map used for data interpretation | |||||||||
 Sample Sample |
| |||||||||
| Biological species | synthetic construct (others) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 6.57 Å | |||||||||
 Authors Authors | Antanasijevic A / Ward AB | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2023 Journal: Proc Natl Acad Sci U S A / Year: 2023Title: Improving the secretion of designed protein assemblies through negative design of cryptic transmembrane domains. Authors: Jing Yang John Wang / Alena Khmelinskaia / William Sheffler / Marcos C Miranda / Aleksandar Antanasijevic / Andrew J Borst / Susana V Torres / Chelsea Shu / Yang Hsia / Una Nattermann / ...Authors: Jing Yang John Wang / Alena Khmelinskaia / William Sheffler / Marcos C Miranda / Aleksandar Antanasijevic / Andrew J Borst / Susana V Torres / Chelsea Shu / Yang Hsia / Una Nattermann / Daniel Ellis / Carl Walkey / Maggie Ahlrichs / Sidney Chan / Alex Kang / Hannah Nguyen / Claire Sydeman / Banumathi Sankaran / Mengyu Wu / Asim K Bera / Lauren Carter / Brooke Fiala / Michael Murphy / David Baker / Andrew B Ward / Neil P King /   Abstract: Computationally designed protein nanoparticles have recently emerged as a promising platform for the development of new vaccines and biologics. For many applications, secretion of designed ...Computationally designed protein nanoparticles have recently emerged as a promising platform for the development of new vaccines and biologics. For many applications, secretion of designed nanoparticles from eukaryotic cells would be advantageous, but in practice, they often secrete poorly. Here we show that designed hydrophobic interfaces that drive nanoparticle assembly are often predicted to form cryptic transmembrane domains, suggesting that interaction with the membrane insertion machinery could limit efficient secretion. We develop a general computational protocol, the Degreaser, to design away cryptic transmembrane domains without sacrificing protein stability. The retroactive application of the Degreaser to previously designed nanoparticle components and nanoparticles considerably improves secretion, and modular integration of the Degreaser into design pipelines results in new nanoparticles that secrete as robustly as naturally occurring protein assemblies. Both the Degreaser protocol and the nanoparticles we describe may be broadly useful in biotechnological applications. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_28862.map.gz emd_28862.map.gz | 114.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-28862-v30.xml emd-28862-v30.xml emd-28862.xml emd-28862.xml | 19.5 KB 19.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_28862_fsc.xml emd_28862_fsc.xml | 11.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_28862.png emd_28862.png | 108.5 KB | ||
| Masks |  emd_28862_msk_1.map emd_28862_msk_1.map | 125 MB |  Mask map Mask map | |
| Others |  emd_28862_half_map_1.map.gz emd_28862_half_map_1.map.gz emd_28862_half_map_2.map.gz emd_28862_half_map_2.map.gz | 93.6 MB 93.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-28862 http://ftp.pdbj.org/pub/emdb/structures/EMD-28862 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28862 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28862 | HTTPS FTP |
-Validation report
| Summary document |  emd_28862_validation.pdf.gz emd_28862_validation.pdf.gz | 721 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_28862_full_validation.pdf.gz emd_28862_full_validation.pdf.gz | 720.6 KB | Display | |
| Data in XML |  emd_28862_validation.xml.gz emd_28862_validation.xml.gz | 18.6 KB | Display | |
| Data in CIF |  emd_28862_validation.cif.gz emd_28862_validation.cif.gz | 24.4 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-28862 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-28862 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-28862 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-28862 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_28862.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_28862.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | KWOCA 4 nanoparticle - The main cryoEM map used for data interpretation | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.15 Å | ||||||||||||||||||||||||||||||||||||
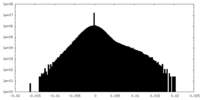
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_28862_msk_1.map emd_28862_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
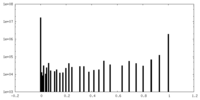
| Density Histograms |
-Half map: KWOCA 4 nanoparticle - Half Map 1
| File | emd_28862_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | KWOCA 4 nanoparticle - Half Map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
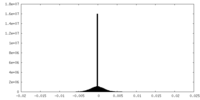
| Density Histograms |
-Half map: KWOCA 4 nanoparticle - Half Map 2
| File | emd_28862_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | KWOCA 4 nanoparticle - Half Map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
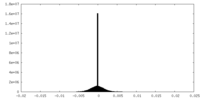
| Density Histograms |
- Sample components
Sample components
-Entire : Designed octahedral nanoparticle KWOCA 4
| Entire | Name: Designed octahedral nanoparticle KWOCA 4 |
|---|---|
| Components |
|
-Supramolecule #1: Designed octahedral nanoparticle KWOCA 4
| Supramolecule | Name: Designed octahedral nanoparticle KWOCA 4 / type: complex / ID: 1 / Chimera: Yes / Parent: 0 / Macromolecule list: all Details: Self-assembling nanoparticle expressed in mammalian cells |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
-Macromolecule #1: KWOCA 4
| Macromolecule | Name: KWOCA 4 / type: protein_or_peptide / ID: 1 / Details: Sequence for expression in mammalian cells / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: METDTLLLWV LLLWVPGSTG DGSHHHHHHG GSEQKLISEE DLSGGGSWSG STEVEKKARE VAKEAVELAS LLRSETAIRV AQAILEAAEA AKRAAEQGKT EVAKLALKVL EEAIELAKEK RSEEALKVVL EIARAALAAA QAAEEGFTDV AKMALEVLER AIELAKDDRS ...String: METDTLLLWV LLLWVPGSTG DGSHHHHHHG GSEQKLISEE DLSGGGSWSG STEVEKKARE VAKEAVELAS LLRSETAIRV AQAILEAAEA AKRAAEQGKT EVAKLALKVL EEAIELAKEK RSEEALKVVL EIARAALAAA QAAEEGFTDV AKMALEVLER AIELAKDDRS EEALKEVLEI ARAALAAAQL AKKGRDDEAR KILMKLRIRI TLRKLEESLR ELRRILEELK EMLERLEKNP DKDVIVKVLK VIVKAIEASV ENQRISAENQ KALAELA |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 3.58 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
Details: TBS + 5% glycerol | ||||||||||||
| Grid | Model: UltrAuFoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Support film - Material: GOLD / Support film - topology: HOLEY / Pretreatment - Type: PLASMA CLEANING / Pretreatment - Time: 10 sec. / Pretreatment - Atmosphere: OTHER | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 283 K / Instrument: FEI VITROBOT MARK IV / Details: Blotting time varied between 3 and 7 seconds.. | ||||||||||||
| Details | Self-assembling nanoparticle was expressed in mammalian cells and purified using affinity chromatography and size-exclusion chromatography. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Frames/image: 1-49 / Number grids imaged: 1 / Number real images: 1341 / Average exposure time: 9.8 sec. / Average electron dose: 50.1 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 1.5 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 36000 |
| Sample stage | Specimen holder model: OTHER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: |
|---|
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)