[English] 日本語
 Yorodumi
Yorodumi- EMDB-28691: Atomic model of the core modifying region of human fatty acid syn... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Atomic model of the core modifying region of human fatty acid synthase in complex with TVB-2640 | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Fatty acid synthase / TRANSFERASE / TRANSFERASE-INHIBITOR complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationfatty-acid synthase system / : / ether lipid biosynthetic process / : / Vitamin B5 (pantothenate) metabolism / : / neutrophil differentiation / enoyl-[acyl-carrier-protein] reductase (NADPH, Re-specific) / : / glandular epithelial cell development ...fatty-acid synthase system / : / ether lipid biosynthetic process / : / Vitamin B5 (pantothenate) metabolism / : / neutrophil differentiation / enoyl-[acyl-carrier-protein] reductase (NADPH, Re-specific) / : / glandular epithelial cell development / : / establishment of endothelial intestinal barrier / glycogen granule / [acyl-carrier-protein] S-acetyltransferase / [acyl-carrier-protein] S-acetyltransferase activity / Fatty acyl-CoA biosynthesis / fatty acyl-[ACP] hydrolase activity / oleoyl-[acyl-carrier-protein] hydrolase / : / modulation by host of viral process / ChREBP activates metabolic gene expression / [acyl-carrier-protein] S-malonyltransferase / [acyl-carrier-protein] S-malonyltransferase activity / 3-hydroxyacyl-[acyl-carrier-protein] dehydratase / beta-ketoacyl-[acyl-carrier-protein] synthase I / NR1H2 & NR1H3 regulate gene expression linked to lipogenesis / mammary gland development / 3-oxoacyl-[acyl-carrier-protein] reductase / 3-oxoacyl-[acyl-carrier-protein] reductase (NADPH) activity / fatty acid synthase activity / phosphopantetheine binding / monocyte differentiation / 3-oxoacyl-[acyl-carrier-protein] synthase activity / cellular response to interleukin-4 / Activation of gene expression by SREBF (SREBP) / fatty acid metabolic process / fatty acid biosynthetic process / osteoblast differentiation / melanosome / cadherin binding / inflammatory response / Golgi apparatus / RNA binding / extracellular exosome / membrane / plasma membrane / cytoplasm / cytosol Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.7 Å | |||||||||
 Authors Authors | Hasan SMN / Keszei A / Mazhab-Jafari MT | |||||||||
| Funding support |  Canada, 2 items Canada, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: Atomic model for core modifying region of human fatty acid synthase in complex with Denifanstat. Authors: S M Naimul Hasan / Jennifer W Lou / Alexander F A Keszei / David L Dai / Mohammad T Mazhab-Jafari /  Abstract: Fatty acid synthase (FASN) catalyzes the de novo synthesis of palmitate, a 16-carbon chain fatty acid that is the primary precursor of lipid metabolism and an important intracellular signaling ...Fatty acid synthase (FASN) catalyzes the de novo synthesis of palmitate, a 16-carbon chain fatty acid that is the primary precursor of lipid metabolism and an important intracellular signaling molecule. FASN is an attractive drug target in diabetes, cancer, fatty liver diseases, and viral infections. Here, we develop an engineered full-length human FASN (hFASN) that enables isolation of the condensing and modifying regions of the protein post-translation. The engineered protein enables electron cryo-microscopy (cryoEM) structure determination of the core modifying region of hFASN to 2.7 Å resolution. Examination of the dehydratase dimer within this region reveals that unlike its close homolog, porcine FASN, the catalytic cavity is close-ended and is accessible only through one opening in the vicinity of the active site. The core modifying region exhibits two major global conformational variabilities that describe long-range bending and twisting motions of the complex in solution. Finally, we solved the structure of this region bound to an anti-cancer drug, Denifanstat (i.e., TVB-2640), demonstrating the utility of our approach as a platform for structure guided design of future hFASN small molecule inhibitors. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_28691.map.gz emd_28691.map.gz | 31 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-28691-v30.xml emd-28691-v30.xml emd-28691.xml emd-28691.xml | 17.6 KB 17.6 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_28691.png emd_28691.png | 72 KB | ||
| Filedesc metadata |  emd-28691.cif.gz emd-28691.cif.gz | 6.9 KB | ||
| Others |  emd_28691_half_map_1.map.gz emd_28691_half_map_1.map.gz emd_28691_half_map_2.map.gz emd_28691_half_map_2.map.gz | 55.3 MB 55.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-28691 http://ftp.pdbj.org/pub/emdb/structures/EMD-28691 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28691 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28691 | HTTPS FTP |
-Validation report
| Summary document |  emd_28691_validation.pdf.gz emd_28691_validation.pdf.gz | 774.7 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_28691_full_validation.pdf.gz emd_28691_full_validation.pdf.gz | 774.3 KB | Display | |
| Data in XML |  emd_28691_validation.xml.gz emd_28691_validation.xml.gz | 12.3 KB | Display | |
| Data in CIF |  emd_28691_validation.cif.gz emd_28691_validation.cif.gz | 14.4 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-28691 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-28691 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-28691 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-28691 | HTTPS FTP |
-Related structure data
| Related structure data |  8eykMC  8eyiC  8gkcC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_28691.map.gz / Format: CCP4 / Size: 59.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_28691.map.gz / Format: CCP4 / Size: 59.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Voxel size | X=Y=Z: 1.03 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_28691_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
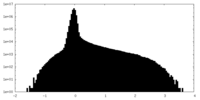
| Projections & Slices |
| ||||||||||||
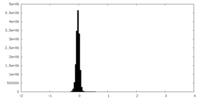
| Density Histograms |
-Half map: #1
| File | emd_28691_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
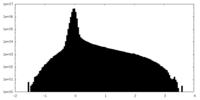
| Projections & Slices |
| ||||||||||||
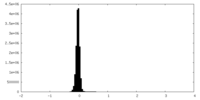
| Density Histograms |
- Sample components
Sample components
-Entire : Fatty acid synthase
| Entire | Name: Fatty acid synthase |
|---|---|
| Components |
|
-Supramolecule #1: Fatty acid synthase
| Supramolecule | Name: Fatty acid synthase / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 340 KDa |
-Macromolecule #1: Fatty acid synthase
| Macromolecule | Name: Fatty acid synthase / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO / EC number: fatty-acid synthase system |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 182.749016 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: GGSPSAAIYN IDTSSESPDH YLVDHTLDGR VLFPATGYLS IVWKTLARAL GLGVEQLPVV FEDVVLHQAT ILPKTGTVSL EVRLLEASR AFEVSENGNL VVSGKVYQWD DPDPRLFDHP ESPTPNPTEP LFLAQAEVYK ELRLRGYDYG PHFQGILEAS L EGDSGRLL ...String: GGSPSAAIYN IDTSSESPDH YLVDHTLDGR VLFPATGYLS IVWKTLARAL GLGVEQLPVV FEDVVLHQAT ILPKTGTVSL EVRLLEASR AFEVSENGNL VVSGKVYQWD DPDPRLFDHP ESPTPNPTEP LFLAQAEVYK ELRLRGYDYG PHFQGILEAS L EGDSGRLL WKDNWVSFMD TMLQMSILGS AKHGLYLPTR VTAIHIDPAT HRQKLYTLQD KAQVADVVVS RWLRVTVAGG VH ISGLHTE SAPRRQQEQQ VPILEKFCFT PHTEEGCLSE RAALQEELQL CKGLVQALQT KVTQQGLKMV VPGLDGAQIP RDP SQQELP RLLSAACRLQ LNGNLQLELA QVLAQERPKL PEDPLLSGLL DSPALKACLD TAVENMPSLK MKVVEVLAGH GHLY SRIPG LLSPHPLLQL SYTATDRHPQ ALEAAQAELQ QHDVAQGQWD PADPAPSALG SADLLVCNCA VAALGDPASA LSNMV AALR EGGFLLLHTL LRGHPLGDIV AFLTSTEPQY GQGILSQDAW ESLFSRVSLR LVGLKKSFYG STLFLCRRPT PQDSPI FLP VDDTSFRWVE SLKGILADED SSRPVWLKAI NCATSGVVGL VNCLRREPGG NRLRCVLLSN LSSTSHVPEV DPGSAEL QK VLQGDLVMNV YRDGAWGAFR HFLLEEDKPE EPTAHAFVST LTRGDLSSIR WVCSSLRHAQ PTCPGAQLCT VYYASLNF R DIMLATGKLS PDAIPGKWTS QDSLLGMEFS GRDASGKRVM GLVPAKGLAT SVLLSPDFLW DVPSNWTLEE AASVPVVYS TAYYALVVRG RVRPGETLLI HSGSGGVGQA AIAIALSLGC RVFTTVGSAE KRAYLQARFP QLDSTSFANS RDTSFEQHVL WHTGGKGVD LVLNSLAEEK LQASVRCLAT HGRFLEIGKF DLSQNHPLGM AIFLKNVTFH GVLLDAFFNE SSADWREVWA L VQAGIRDG VVRPLKCTVF HGAQVEDAFR YMAQGKHIGK VVVQVLAEEP EAVLKGAKPK LMSAISKTFC PAHKSYIIAG GL GGFGLEL AQWLIQRGVQ KLVLTSRSGI RTGYQAKQVR RWRRQGVQVQ VSTSNISSLE GARGLIAEAA QLGPVGGVFN LAV VLRDGL LENQTPEFFQ DVCKPKYSGT LNLDRVTREA CPELDYFVVF SSVSCGRGNA GQSNYGFANS AMERICEKRR HEGL PGLAV QWGAIGDVGI LVETMSTNDT IVSGTLPQRM ASCLEVLDLF LNQPHMVLSS FVLAEKAAAY RDRDSQRDLV EAVAH ILGI RDLAAVNLDS SLADLGLDSL MSVEVRQTLE RELNLVLSVR EVRQLTLRKL QELSSKADEA SELACPTPKE DGLAQQ QTQ LNLRSLLVNP EGPTLMRLNS VQSSERPLFL VHPIEGSTTV FHSLASRLSI PTYGLQCTRA APLDSIHSLA AYYIDCI RQ VQPEGPYRVA GYSYGACVAF EMCSQLQAQQ SPAPTHNSLF LFDGSPTYVL AYTQSYRAKL TPGCEAEAET EAICFFVQ Q FTDMEHNRVL EALLPLKGLE ERVAAAVDLI IKSHQGLDRQ ELSFAARSFY YKLRAAEQYT PKAKYHGNVM LLRAKTGGA YGEDLGADYN LSQVCDGKVS VHVIEGDHRT LLEGSGLESI ISIIHSSLAE PRVSVREGLE SRGPHHHHHH UniProtKB: Fatty acid synthase |
-Macromolecule #2: denifanstat
| Macromolecule | Name: denifanstat / type: ligand / ID: 2 / Number of copies: 2 / Formula: X5O |
|---|---|
| Molecular weight | Theoretical: 439.552 Da |
| Chemical component information | 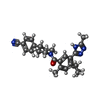 ChemComp-X5O: |
-Macromolecule #3: NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE
| Macromolecule | Name: NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE type: ligand / ID: 3 / Number of copies: 4 / Formula: NDP |
|---|---|
| Molecular weight | Theoretical: 745.421 Da |
| Chemical component information |  ChemComp-NDP: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.8 mg/mL |
|---|---|
| Buffer | pH: 7.4 |
| Grid | Model: Homemade / Material: COPPER/RHODIUM / Mesh: 400 / Support film - Material: GOLD / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 25 sec. |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Number grids imaged: 1 / Number real images: 4801 / Average electron dose: 50.76 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.6 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: OTHER |
|---|---|
| Final reconstruction | Applied symmetry - Point group: C1 (asymmetric) / Resolution.type: BY AUTHOR / Resolution: 2.7 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: cryoSPARC (ver. 3.3) / Number images used: 311983 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD / Software - Name: cryoSPARC (ver. 3.3) |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD / Software - Name: cryoSPARC (ver. 3.3) |
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT |
|---|---|
| Output model |  PDB-8eyk: |
 Movie
Movie Controller
Controller











 Z
Z Y
Y X
X