+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | DH272.2 Fab in Complex with CH848 TF DS SOSIP Env | |||||||||
 Map data Map data | DH272.2 Fab bound to CH848 TF DS SOSIP Env | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /   Human immunodeficiency virus Human immunodeficiency virus | |||||||||
| Method | single particle reconstruction / negative staining / Resolution: 14.35 Å | |||||||||
 Authors Authors | Fera D | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Front Immunol / Year: 2022 Journal: Front Immunol / Year: 2022Title: Analysis of two cooperating antibodies unveils immune pressure imposed on HIV Env to elicit a V3-glycan supersite broadly neutralizing antibody lineage. Authors: Maxwell T Finkelstein / Emma Parker Miller / Molly C Erdman / Daniela Fera /  Abstract: Elicitation of broadly neutralizing antibodies (bnAbs) is a goal of vaccine design as a strategy for targeting highly divergent strains of HIV-1. Current HIV-1 vaccine design efforts seek to elicit ...Elicitation of broadly neutralizing antibodies (bnAbs) is a goal of vaccine design as a strategy for targeting highly divergent strains of HIV-1. Current HIV-1 vaccine design efforts seek to elicit bnAbs by first eliciting their precursors through prime-boost regimens. This requires an understanding of the co-evolution between viruses and antibodies. Towards this goal, we have analyzed two cooperating antibodies, DH475 and DH272, which exerted pressure on the HIV population in an infected donor, called CH848, to evolve in such a way that it became sensitive to the V3-glycan supersite DH270 bnAb lineage. We obtained a 2.90Å crystal structure of DH475 in complex with the Man glycan and a negative stain EM model of DH272 in complex with the HIV-1 spike trimer, Env. Coupled with additional modeling studies and biochemical data, our studies reveal that DH475 contacts a V3- and V4-glycan dependent epitope accessible on an open or shed Env and that DH272 makes critical contacts with the V1V2 and V3 loops on HIV-1 Env. Using these data, we suggest a prime-boost regimen that may facilitate the initiation of DH270-like bnAb precursors. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_28120.map.gz emd_28120.map.gz | 14.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-28120-v30.xml emd-28120-v30.xml emd-28120.xml emd-28120.xml | 12.6 KB 12.6 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_28120.png emd_28120.png | 13.8 KB | ||
| Others |  emd_28120_half_map_1.map.gz emd_28120_half_map_1.map.gz emd_28120_half_map_2.map.gz emd_28120_half_map_2.map.gz | 26.6 MB 26.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-28120 http://ftp.pdbj.org/pub/emdb/structures/EMD-28120 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28120 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28120 | HTTPS FTP |
-Validation report
| Summary document |  emd_28120_validation.pdf.gz emd_28120_validation.pdf.gz | 690.3 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_28120_full_validation.pdf.gz emd_28120_full_validation.pdf.gz | 689.9 KB | Display | |
| Data in XML |  emd_28120_validation.xml.gz emd_28120_validation.xml.gz | 10.3 KB | Display | |
| Data in CIF |  emd_28120_validation.cif.gz emd_28120_validation.cif.gz | 11.9 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-28120 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-28120 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-28120 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-28120 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_28120.map.gz / Format: CCP4 / Size: 28.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_28120.map.gz / Format: CCP4 / Size: 28.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | DH272.2 Fab bound to CH848 TF DS SOSIP Env | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 3 Å | ||||||||||||||||||||||||||||||||||||
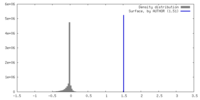
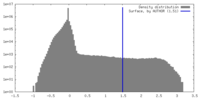
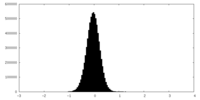
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: DH272.2 Fab bound to CH848 TF DS SOSIP Env
| File | emd_28120_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | DH272.2 Fab bound to CH848 TF DS SOSIP Env | ||||||||||||
| Projections & Slices |
| ||||||||||||
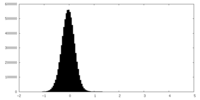
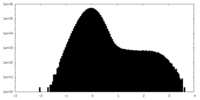
| Density Histograms |
-Half map: DH272.2 Fab bound to CH848 TF DS SOSIP Env
| File | emd_28120_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | DH272.2 Fab bound to CH848 TF DS SOSIP Env | ||||||||||||
| Projections & Slices |
| ||||||||||||
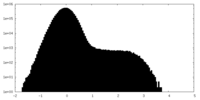
| Density Histograms |
- Sample components
Sample components
-Entire : Complex between DH272.2 Fab and CH848 TF DS SOSIP Env
| Entire | Name: Complex between DH272.2 Fab and CH848 TF DS SOSIP Env |
|---|---|
| Components |
|
-Supramolecule #1: Complex between DH272.2 Fab and CH848 TF DS SOSIP Env
| Supramolecule | Name: Complex between DH272.2 Fab and CH848 TF DS SOSIP Env / type: complex / Chimera: Yes / ID: 1 / Parent: 0 |
|---|
-Supramolecule #2: DH272.2 Fab
| Supramolecule | Name: DH272.2 Fab / type: complex / Chimera: Yes / ID: 2 / Parent: 1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  Homo sapiens (human) / Recombinant cell: HEK-293T Homo sapiens (human) / Recombinant cell: HEK-293T |
-Supramolecule #3: CH848 TF DS SOSIP Env
| Supramolecule | Name: CH848 TF DS SOSIP Env / type: complex / Chimera: Yes / ID: 3 / Parent: 1 |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus Human immunodeficiency virus |
| Recombinant expression | Organism:  Homo sapiens (human) / Recombinant cell: HEK293S GnTi- Homo sapiens (human) / Recombinant cell: HEK293S GnTi- |
-Experimental details
-Structure determination
| Method | negative staining |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Staining | Type: NEGATIVE / Material: Uranyl Formate |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI F20 |
|---|---|
| Image recording | Film or detector model: OTHER / Average electron dose: 25.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 3.0 µm / Nominal defocus min: 3.0 µm |
| Experimental equipment |  Model: Tecnai F20 / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 14.35 Å / Resolution method: FSC 0.5 CUT-OFF / Number images used: 19124 |
|---|---|
| Initial angle assignment | Type: NOT APPLICABLE |
| Final angle assignment | Type: NOT APPLICABLE |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)