+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | 9H2 Fab-Sabin poliovirus 3 complex | ||||||||||||
 Map data Map data | Cryosparc sharpened | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | Complex / Fab / poliovirus / neutralizing / VIRUS-IMMUNE SYSTEM complex | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationcaveolin-mediated endocytosis of virus by host cell / symbiont-mediated suppression of host cytoplasmic pattern recognition receptor signaling pathway via inhibition of RIG-I activity / symbiont-mediated suppression of host cytoplasmic pattern recognition receptor signaling pathway via inhibition of MDA-5 activity / picornain 2A / symbiont-mediated suppression of host mRNA export from nucleus / symbiont-mediated suppression of host cytoplasmic pattern recognition receptor signaling pathway via inhibition of MAVS activity / symbiont genome entry into host cell via pore formation in plasma membrane / picornain 3C / ribonucleoside triphosphate phosphatase activity / T=pseudo3 icosahedral viral capsid ...caveolin-mediated endocytosis of virus by host cell / symbiont-mediated suppression of host cytoplasmic pattern recognition receptor signaling pathway via inhibition of RIG-I activity / symbiont-mediated suppression of host cytoplasmic pattern recognition receptor signaling pathway via inhibition of MDA-5 activity / picornain 2A / symbiont-mediated suppression of host mRNA export from nucleus / symbiont-mediated suppression of host cytoplasmic pattern recognition receptor signaling pathway via inhibition of MAVS activity / symbiont genome entry into host cell via pore formation in plasma membrane / picornain 3C / ribonucleoside triphosphate phosphatase activity / T=pseudo3 icosahedral viral capsid / host cell cytoplasmic vesicle membrane / cytoplasmic vesicle membrane / endocytosis involved in viral entry into host cell / nucleoside-triphosphate phosphatase / channel activity / monoatomic ion transmembrane transport / DNA replication / RNA helicase activity / symbiont-mediated suppression of host innate immune response / induction by virus of host autophagy / symbiont entry into host cell / RNA-directed RNA polymerase / viral RNA genome replication / cysteine-type endopeptidase activity / RNA-dependent RNA polymerase activity / virus-mediated perturbation of host defense response / DNA-templated transcription / host cell nucleus / virion attachment to host cell / structural molecule activity / proteolysis / RNA binding / ATP binding / membrane / metal ion binding Similarity search - Function | ||||||||||||
| Biological species |  Human poliovirus 3 strain Sabin / Human poliovirus 3 strain Sabin /  Homo sapiens (human) Homo sapiens (human) | ||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.91 Å | ||||||||||||
 Authors Authors | Charnesky AJ | ||||||||||||
| Funding support |  United States, 3 items United States, 3 items
| ||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: A human monoclonal antibody binds within the poliovirus receptor-binding site to neutralize all three serotypes. Authors: Andrew J Charnesky / Julia E Faust / Hyunwook Lee / Rama Devudu Puligedda / Daniel J Goetschius / Nadia M DiNunno / Vaskar Thapa / Carol M Bator / Sung Hyun Joseph Cho / Rahnuma Wahid / ...Authors: Andrew J Charnesky / Julia E Faust / Hyunwook Lee / Rama Devudu Puligedda / Daniel J Goetschius / Nadia M DiNunno / Vaskar Thapa / Carol M Bator / Sung Hyun Joseph Cho / Rahnuma Wahid / Kutub Mahmood / Scott Dessain / Konstantin M Chumakov / Amy Rosenfeld / Susan L Hafenstein /  Abstract: Global eradication of poliovirus remains elusive, and it is critical to develop next generation vaccines and antivirals. In support of this goal, we map the epitope of human monoclonal antibody 9H2 ...Global eradication of poliovirus remains elusive, and it is critical to develop next generation vaccines and antivirals. In support of this goal, we map the epitope of human monoclonal antibody 9H2 which is able to neutralize the three serotypes of poliovirus. Using cryo-EM we solve the near-atomic structures of 9H2 fragments (Fab) bound to capsids of poliovirus serotypes 1, 2, and 3. The Fab-virus complexes show that Fab interacts with the same binding mode for each serotype and at the same angle of interaction relative to the capsid surface. For each of the Fab-virus complexes, we find that the binding site overlaps with the poliovirus receptor (PVR) binding site and maps across and into a depression in the capsid called the canyon. No conformational changes to the capsid are induced by Fab binding for any complex. Competition binding experiments between 9H2 and PVR reveal that 9H2 impedes receptor binding. Thus, 9H2 outcompetes the receptor to neutralize poliovirus. The ability to neutralize all three serotypes, coupled with the critical importance of the conserved receptor binding site make 9H2 an attractive antiviral candidate for future development. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_27949.map.gz emd_27949.map.gz | 631.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-27949-v30.xml emd-27949-v30.xml emd-27949.xml emd-27949.xml | 23.3 KB 23.3 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_27949.png emd_27949.png | 286.7 KB | ||
| Filedesc metadata |  emd-27949.cif.gz emd-27949.cif.gz | 6.7 KB | ||
| Others |  emd_27949_additional_1.map.gz emd_27949_additional_1.map.gz emd_27949_half_map_1.map.gz emd_27949_half_map_1.map.gz emd_27949_half_map_2.map.gz emd_27949_half_map_2.map.gz | 589.5 MB 613.5 MB 613.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-27949 http://ftp.pdbj.org/pub/emdb/structures/EMD-27949 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27949 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27949 | HTTPS FTP |
-Validation report
| Summary document |  emd_27949_validation.pdf.gz emd_27949_validation.pdf.gz | 939.8 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_27949_full_validation.pdf.gz emd_27949_full_validation.pdf.gz | 939.4 KB | Display | |
| Data in XML |  emd_27949_validation.xml.gz emd_27949_validation.xml.gz | 20.4 KB | Display | |
| Data in CIF |  emd_27949_validation.cif.gz emd_27949_validation.cif.gz | 24.1 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27949 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27949 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27949 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27949 | HTTPS FTP |
-Related structure data
| Related structure data |  8e8xMC  8e8lC  8e8rC  8e8sC  8e8yC  8e8zC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_27949.map.gz / Format: CCP4 / Size: 669.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_27949.map.gz / Format: CCP4 / Size: 669.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryosparc sharpened | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.1 Å | ||||||||||||||||||||||||||||||||||||
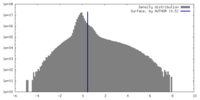
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: DeepEMhancer sharpened
| File | emd_27949_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | DeepEMhancer sharpened | ||||||||||||
| Projections & Slices |
| ||||||||||||
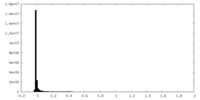
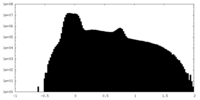
| Density Histograms |
-Half map: Half Map A
| File | emd_27949_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half Map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
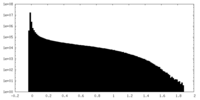
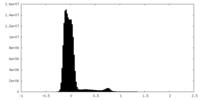
| Density Histograms |
-Half map: Half Map B
| File | emd_27949_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half Map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
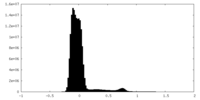
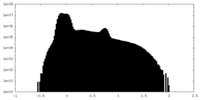
| Density Histograms |
- Sample components
Sample components
-Entire : Human poliovirus 3 strain Sabin
| Entire | Name:  Human poliovirus 3 strain Sabin Human poliovirus 3 strain Sabin |
|---|---|
| Components |
|
-Supramolecule #1: Human poliovirus 3 strain Sabin
| Supramolecule | Name: Human poliovirus 3 strain Sabin / type: virus / ID: 1 / Parent: 0 / Macromolecule list: #1-#6 / NCBI-ID: 270338 / Sci species name: Human poliovirus 3 strain Sabin / Virus type: VIRION / Virus isolate: SEROTYPE / Virus enveloped: No / Virus empty: No |
|---|---|
| Host (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Capsid protein VP1
| Macromolecule | Name: Capsid protein VP1 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Human poliovirus 3 strain Sabin / Strain: Sabin Human poliovirus 3 strain Sabin / Strain: Sabin |
| Molecular weight | Theoretical: 31.358221 KDa |
| Sequence | String: QDSLPDTKAS GPAHSKEVPA LTAVETGATN PLAPSDTVQT RHVVQRRSRS ESTIESFFAR GACVAIIEVD NEQPTTRAQK LFAMWRITY KDTVQLRRKL EFFTYSRFDM EFTFVVTANF TNANNGHALN QVYQIMYIPP GAPTPKSWDD YTWQTSSNPS I FYTYGAAP ...String: QDSLPDTKAS GPAHSKEVPA LTAVETGATN PLAPSDTVQT RHVVQRRSRS ESTIESFFAR GACVAIIEVD NEQPTTRAQK LFAMWRITY KDTVQLRRKL EFFTYSRFDM EFTFVVTANF TNANNGHALN QVYQIMYIPP GAPTPKSWDD YTWQTSSNPS I FYTYGAAP ARISVPYVGL ANAYSHFYDG FAKVPLKTDA NDQIGDSLYS AMTVDDFGVL AVRVVNDHNP TKVTSKVRIY MK PKHVRVW CPRPPRAVPY YGPGVDYRNN LDPLSEKGLT TY UniProtKB: Genome polyprotein |
-Macromolecule #2: Capsid protein VP2
| Macromolecule | Name: Capsid protein VP2 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Human poliovirus 3 strain Sabin / Strain: Sabin Human poliovirus 3 strain Sabin / Strain: Sabin |
| Molecular weight | Theoretical: 29.412111 KDa |
| Sequence | String: YSDRVLQLTL GNSTITTQEA ANSVVAYGRW PEFIRDDEAN PVDQPTEPDV ATCRFYTLDT VMWGKESKGW WWKLPDALRD MGLFGQNMY YHYLGRSGYT VHVQCNASKF HQGALGVFAI PEYCLAGDSD KQRYTSYANA NPGERGGKFY SQFNKDNAVT S PKREFCPV ...String: YSDRVLQLTL GNSTITTQEA ANSVVAYGRW PEFIRDDEAN PVDQPTEPDV ATCRFYTLDT VMWGKESKGW WWKLPDALRD MGLFGQNMY YHYLGRSGYT VHVQCNASKF HQGALGVFAI PEYCLAGDSD KQRYTSYANA NPGERGGKFY SQFNKDNAVT S PKREFCPV DYLLGCGVLL GNAFVYPHQI INLRTNNSAT IVLPYVNALA IDSMVKHNNW GIAILPLSPL DFAQDSSVEI PI TVTIAPM CSEFNGLRNV TAPKFQ UniProtKB: Genome polyprotein |
-Macromolecule #3: Capsid protein VP3
| Macromolecule | Name: Capsid protein VP3 / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Human poliovirus 3 strain Sabin / Strain: Sabin Human poliovirus 3 strain Sabin / Strain: Sabin |
| Molecular weight | Theoretical: 25.950723 KDa |
| Sequence | String: GLPVLNTPGS NQYLTSDNHQ SPCAIPEFDV TPPIDIPGEV KNMMELAEID TMIPLNLEST KRNTMDMYRV TLSDSADLSQ PILCLSLSP AFDPRLSHTM LGEVLNYYTH WAGSLKFTFL FCGSMMATGK ILVAYAPPGA QPPTSRKEAM LGTHVIWDLG L QSSCTMVV ...String: GLPVLNTPGS NQYLTSDNHQ SPCAIPEFDV TPPIDIPGEV KNMMELAEID TMIPLNLEST KRNTMDMYRV TLSDSADLSQ PILCLSLSP AFDPRLSHTM LGEVLNYYTH WAGSLKFTFL FCGSMMATGK ILVAYAPPGA QPPTSRKEAM LGTHVIWDLG L QSSCTMVV PWISNVTYRQ TTQDSFTEGG YISMFYQTRI VVPLSTPKSM SMLGFVSACN DFSVRLLRDT THISQSA UniProtKB: Genome polyprotein |
-Macromolecule #4: Capsid protein VP4
| Macromolecule | Name: Capsid protein VP4 / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Human poliovirus 3 strain Sabin / Strain: Sabin Human poliovirus 3 strain Sabin / Strain: Sabin |
| Molecular weight | Theoretical: 7.320917 KDa |
| Sequence | String: GAQVSSQKVG AHENSNRAYG GSTINYTTIN YYKDSASNAA SKQDYSQDPS KFTEPLKDVL IKTAPALN UniProtKB: Genome polyprotein |
-Macromolecule #5: 9H2 Fab heavy chain
| Macromolecule | Name: 9H2 Fab heavy chain / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 14.052716 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: LVQSGAELKK PGASVKFSCQ ASGFTFTTYD IHWVRQAPGQ GLEWMGMISP SRDSTIYAQK FQGRVTMTSD TSTSTVYMEL TSLRSEDTA LYYCATASRP SAWVFRSLYT YYYMDVWGTG TTVTVSS |
-Macromolecule #6: 9H2 Fab light chain
| Macromolecule | Name: 9H2 Fab light chain / type: protein_or_peptide / ID: 6 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 11.738903 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QSALTQPASV SGSPGQSITI SCTGTITDIG YYNYVSWYQQ HPGKAPKLII FDVTNRPSGV SDRFSGSKSG NTASLTISGL QAEDEGDYY CFSHRSNNIR VFGGGTKLTV L |
-Macromolecule #7: PALMITIC ACID
| Macromolecule | Name: PALMITIC ACID / type: ligand / ID: 7 / Number of copies: 1 / Formula: PLM |
|---|---|
| Molecular weight | Theoretical: 256.424 Da |
| Chemical component information |  ChemComp-PLM: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: INSILICO MODEL |
|---|---|
| Final reconstruction | Applied symmetry - Point group: I (icosahedral) / Resolution.type: BY AUTHOR / Resolution: 2.91 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 52115 |
| Initial angle assignment | Type: RANDOM ASSIGNMENT |
| Final angle assignment | Type: PROJECTION MATCHING |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)