[English] 日本語
 Yorodumi
Yorodumi- EMDB-26507: SARS-CoV-2 spike in complex with Multivalent miniprotein inhibito... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | SARS-CoV-2 spike in complex with Multivalent miniprotein inhibitor FUS231-P24 (2RBDs open) | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.9 Å | |||||||||
 Authors Authors | Park YJ / Seattle Structural Genomics Center for Infectious Disease (SSGCID) / Veesler D | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Sci Transl Med / Year: 2022 Journal: Sci Transl Med / Year: 2022Title: Multivalent designed proteins neutralize SARS-CoV-2 variants of concern and confer protection against infection in mice. Authors: Andrew C Hunt / James Brett Case / Young-Jun Park / Longxing Cao / Kejia Wu / Alexandra C Walls / Zhuoming Liu / John E Bowen / Hsien-Wei Yeh / Shally Saini / Louisa Helms / Yan Ting Zhao / ...Authors: Andrew C Hunt / James Brett Case / Young-Jun Park / Longxing Cao / Kejia Wu / Alexandra C Walls / Zhuoming Liu / John E Bowen / Hsien-Wei Yeh / Shally Saini / Louisa Helms / Yan Ting Zhao / Tien-Ying Hsiang / Tyler N Starr / Inna Goreshnik / Lisa Kozodoy / Lauren Carter / Rashmi Ravichandran / Lydia B Green / Wadim L Matochko / Christy A Thomson / Bastian Vögeli / Antje Krüger / Laura A VanBlargan / Rita E Chen / Baoling Ying / Adam L Bailey / Natasha M Kafai / Scott E Boyken / Ajasja Ljubetič / Natasha Edman / George Ueda / Cameron M Chow / Max Johnson / Amin Addetia / Mary-Jane Navarro / Nuttada Panpradist / Michael Gale / Benjamin S Freedman / Jesse D Bloom / Hannele Ruohola-Baker / Sean P J Whelan / Lance Stewart / Michael S Diamond / David Veesler / Michael C Jewett / David Baker /    Abstract: New variants of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) continue to arise and prolong the coronavirus disease 2019 (COVID-19) pandemic. Here, we used a cell-free expression ...New variants of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) continue to arise and prolong the coronavirus disease 2019 (COVID-19) pandemic. Here, we used a cell-free expression workflow to rapidly screen and optimize constructs containing multiple computationally designed miniprotein inhibitors of SARS-CoV-2. We found the broadest efficacy was achieved with a homotrimeric version of the 75-residue angiotensin-converting enzyme 2 (ACE2) mimic AHB2 (TRI2-2) designed to geometrically match the trimeric spike architecture. Consistent with the design model, in the cryo-electron microscopy structure TRI2-2 forms a tripod at the apex of the spike protein that engaged all three receptor binding domains simultaneously. TRI2-2 neutralized Omicron (B.1.1.529), Delta (B.1.617.2), and all other variants tested with greater potency than the monoclonal antibodies used clinically for the treatment of COVID-19. TRI2-2 also conferred prophylactic and therapeutic protection against SARS-CoV-2 challenge when administered intranasally in mice. Designed miniprotein receptor mimics geometrically arrayed to match pathogen receptor binding sites could be a widely applicable antiviral therapeutic strategy with advantages over antibodies in greater resistance to viral escape and antigenic drift, and advantages over native receptor traps in lower chances of autoimmune responses. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_26507.map.gz emd_26507.map.gz | 230.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-26507-v30.xml emd-26507-v30.xml emd-26507.xml emd-26507.xml | 22.4 KB 22.4 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_26507.png emd_26507.png | 48.4 KB | ||
| Others |  emd_26507_additional_1.map.gz emd_26507_additional_1.map.gz emd_26507_half_map_1.map.gz emd_26507_half_map_1.map.gz emd_26507_half_map_2.map.gz emd_26507_half_map_2.map.gz | 122.5 MB 226.4 MB 226.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-26507 http://ftp.pdbj.org/pub/emdb/structures/EMD-26507 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26507 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26507 | HTTPS FTP |
-Validation report
| Summary document |  emd_26507_validation.pdf.gz emd_26507_validation.pdf.gz | 784.1 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_26507_full_validation.pdf.gz emd_26507_full_validation.pdf.gz | 783.7 KB | Display | |
| Data in XML |  emd_26507_validation.xml.gz emd_26507_validation.xml.gz | 16 KB | Display | |
| Data in CIF |  emd_26507_validation.cif.gz emd_26507_validation.cif.gz | 19 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26507 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26507 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26507 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26507 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_26507.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_26507.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.16 Å | ||||||||||||||||||||||||||||||||||||
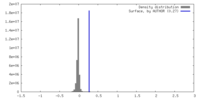
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: #1
| File | emd_26507_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
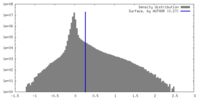
| Density Histograms |
-Half map: #1
| File | emd_26507_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
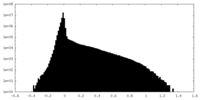
| Density Histograms |
-Half map: #2
| File | emd_26507_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
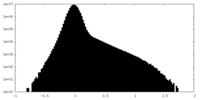
| Density Histograms |
- Sample components
Sample components
-Entire : SARS-CoV-2 spike in complex with multivalent minibinder FUS231-P2...
| Entire | Name: SARS-CoV-2 spike in complex with multivalent minibinder FUS231-P24 (2RBDs open) |
|---|---|
| Components |
|
-Supramolecule #1: SARS-CoV-2 spike in complex with multivalent minibinder FUS231-P2...
| Supramolecule | Name: SARS-CoV-2 spike in complex with multivalent minibinder FUS231-P24 (2RBDs open) type: complex / Chimera: Yes / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
-Supramolecule #2: SARS-CoV-2 Hexapro spike protein
| Supramolecule | Name: SARS-CoV-2 Hexapro spike protein / type: complex / Chimera: Yes / ID: 2 / Parent: 1 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Supramolecule #3: Multivalent miniprotein inhibitor FUS231-P24
| Supramolecule | Name: Multivalent miniprotein inhibitor FUS231-P24 / type: complex / Chimera: Yes / ID: 3 / Parent: 1 / Macromolecule list: #2 |
|---|---|
| Source (natural) | Organism: Synthetic construct (others) |
| Recombinant expression | Organism:  |
-Macromolecule #1: SARS-CoV-2 Hexapro Spike protein
| Macromolecule | Name: SARS-CoV-2 Hexapro Spike protein / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MFVFLVLLPL VSSQCVNLTT RTQLPPAYTN SFTRGVYYPD KVFRSSVLHS TQDLFLPFFS NVTWFHAIHV SGTNGTKRFD NPVLPFNDGV YFASTEKSNI IRGWIFGTTL DSKTQSLLIV NNATNVVIKV CEFQFCNDPF LGVYYHKNNK SWMESEFRVY SSANNCTFEY ...String: MFVFLVLLPL VSSQCVNLTT RTQLPPAYTN SFTRGVYYPD KVFRSSVLHS TQDLFLPFFS NVTWFHAIHV SGTNGTKRFD NPVLPFNDGV YFASTEKSNI IRGWIFGTTL DSKTQSLLIV NNATNVVIKV CEFQFCNDPF LGVYYHKNNK SWMESEFRVY SSANNCTFEY VSQPFLMDLE GKQGNFKNLR EFVFKNIDGY FKIYSKHTPI NLVRDLPQGF SALEPLVDLP IGINITRFQT LLALHRSYLT PGDSSSGWTA GAAAYYVGYL QPRTFLLKYN ENGTITDAVD CALDPLSETK CTLKSFTVEK GIYQTSNFRV QPTESIVRFP NITNLCPFGE VFNATRFASV YAWNRKRISN CVADYSVLYN SASFSTFKCY GVSPTKLNDL CFTNVYADSF VIRGDEVRQI APGQTGKIAD YNYKLPDDFT GCVIAWNSNN LDSKVGGNYN YLYRLFRKSN LKPFERDIST EIYQAGSTPC NGVEGFNCYF PLQSYGFQPT NGVGYQPYRV VVLSFELLHA PATVCGPKKS TNLVKNKCVN FNFNGLTGTG VLTESNKKFL PFQQFGRDIA DTTDAVRDPQ TLEILDITPC SFGGVSVITP GTNTSNQVAV LYQDVNCTEV PVAIHADQLT PTWRVYSTGS NVFQTRAGCL IGAEHVNNSY ECDIPIGAGI CASYQTQTNS PGSASSVASQ SIIAYTMSLG AENSVAYSNN SIAIPTNFTI SVTTEILPVS MTKTSVDCTM YICGDSTECS NLLLQYGSFC TQLNRALTGI AVEQDKNTQE VFAQVKQIYK TPPIKDFGGF NFSQILPDPS KPSKRSPIED LLFNKVTLAD AGFIKQYGDC LGDIAARDLI CAQKFNGLTV LPPLLTDEMI AQYTSALLAG TITSGWTFGA GPALQIPFPM QMAYRFNGIG VTQNVLYENQ KLIANQFNSA IGKIQDSLSS TPSALGKLQD VVNQNAQALN TLVKQLSSNF GAISSVLNDI LSRLDPPEAE VQIDRLITGR LQSLQTYVTQ QLIRAAEIRA SANLAATKMS ECVLGQSKRV DFCGKGYHLM SFPQSAPHGV VFLHVTYVPA QEKNFTTAPA ICHDGKAHFP REGVFVSNGT HWFVTQRNFY EPQIITTDNT FVSGNCDVVI GIVNNTVYDP LQPELDSFKE ELDKYFKNHT SPDVDLGDIS GINASVVNIQ KEIDRLNEVA KNLNESLIDL QELGKYEQGS GYIPEAPRDG QAYVRKDGEW VLLSTFLGRS LEVLFQGPGH HHHHHHHSAW SHPQFEKGGG SGGGGSGGSA WSHPQFEK |
-Macromolecule #2: Multivalent miniprotein inhibitor FUS231-P24
| Macromolecule | Name: Multivalent miniprotein inhibitor FUS231-P24 / type: protein_or_peptide / ID: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: Synthetic construct (others) |
| Recombinant expression | Organism:  |
| Sequence | String: MEKKI NLDE LHMQMTDLVY EALHFAKDEE FQKHVFQLFE KATKAYKNKD RQKLEKVVEE LKELLERLLS GGASPAAPA PASPAAPAPS APAGG ELEE QVMHVLDQVS ELAHELLHKL TGEELERAAY FNWWATEMML ELIKSDDERE IREIEEEAAR ILEHLEELAR T ...String: MEKKI NLDE LHMQMTDLVY EALHFAKDEE FQKHVFQLFE KATKAYKNKD RQKLEKVVEE LKELLERLLS GGASPAAPA PASPAAPAPS APAGG ELEE QVMHVLDQVS ELAHELLHKL TGEELERAAY FNWWATEMML ELIKSDDERE IREIEEEAAR ILEHLEELAR T GGASPAAP APASPAAPAP SAPAGG DKE NVLQKIYEIM KELERLGHAE ASMQVSDLIY EFMKTKDENL LEEAERLLEE VKR GGSGSS GSAWSHPQFE KGGGSGGGSG GSAWSHPQFE K |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS GLACIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.5 µm |
- Image processing
Image processing
| Startup model | Type of model: OTHER / Details: cryoSPARC ab initio |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.9 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: cryoSPARC / Number images used: 112075 |
| Initial angle assignment | Type: PROJECTION MATCHING |
| Final angle assignment | Type: PROJECTION MATCHING |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)