[English] 日本語
 Yorodumi
Yorodumi- EMDB-2637: Negative stain electron microscopy of Bacillus subtilis RNA polym... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-2637 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Negative stain electron microscopy of Bacillus subtilis RNA polymerase with YkzG-GFP fusion | |||||||||
 Map data Map data | Reconstruction of B. subtilis RNAP with YkzG-GFP fusion | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | RNA polymerase / small subunit / transcription / Bacillus subtilis | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / negative staining / Resolution: 24.0 Å | |||||||||
 Authors Authors | Keller A / Yang X / Korelusova J / Delumeau O / Krasny L / Lewis PJ | |||||||||
 Citation Citation |  Journal: J Bacteriol / Year: 2014 Journal: J Bacteriol / Year: 2014Title: ε, a new subunit of RNA polymerase found in gram-positive bacteria. Authors: Andrew N Keller / Xiao Yang / Jana Wiedermannová / Olivier Delumeau / Libor Krásný / Peter J Lewis /    Abstract: RNA polymerase in bacteria is a multisubunit protein complex that is essential for gene expression. We have identified a new subunit of RNA polymerase present in the high-A+T Firmicutes phylum of ...RNA polymerase in bacteria is a multisubunit protein complex that is essential for gene expression. We have identified a new subunit of RNA polymerase present in the high-A+T Firmicutes phylum of Gram-positive bacteria and have named it ε. Previously ε had been identified as a small protein (ω1) that copurified with RNA polymerase. We have solved the structure of ε by X-ray crystallography and show that it is not an ω subunit. Rather, ε bears remarkable similarity to the Gp2 family of phage proteins involved in the inhibition of host cell transcription following infection. Deletion of ε shows no phenotype and has no effect on the transcriptional profile of the cell. Determination of the location of ε within the assembly of RNA polymerase core by single-particle analysis suggests that it binds toward the downstream side of the DNA binding cleft. Due to the structural similarity of ε with Gp2 and the fact they bind similar regions of RNA polymerase, we hypothesize that ε may serve a role in protection from phage infection. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_2637.map.gz emd_2637.map.gz | 1.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-2637-v30.xml emd-2637-v30.xml emd-2637.xml emd-2637.xml | 7.7 KB 7.7 KB | Display Display |  EMDB header EMDB header |
| Images |  EMD-2637.png EMD-2637.png emd-2637.png emd-2637.png | 131 KB 131 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-2637 http://ftp.pdbj.org/pub/emdb/structures/EMD-2637 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-2637 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-2637 | HTTPS FTP |
-Validation report
| Summary document |  emd_2637_validation.pdf.gz emd_2637_validation.pdf.gz | 211.3 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_2637_full_validation.pdf.gz emd_2637_full_validation.pdf.gz | 210.5 KB | Display | |
| Data in XML |  emd_2637_validation.xml.gz emd_2637_validation.xml.gz | 5.2 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-2637 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-2637 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-2637 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-2637 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_2637.map.gz / Format: CCP4 / Size: 1.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_2637.map.gz / Format: CCP4 / Size: 1.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Reconstruction of B. subtilis RNAP with YkzG-GFP fusion | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 3.6 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
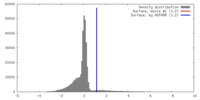
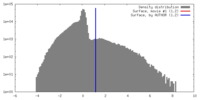
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : RNA polymerase YkzG-GFP
| Entire | Name: RNA polymerase YkzG-GFP |
|---|---|
| Components |
|
-Supramolecule #1000: RNA polymerase YkzG-GFP
| Supramolecule | Name: RNA polymerase YkzG-GFP / type: sample / ID: 1000 / Number unique components: 1 |
|---|---|
| Molecular weight | Experimental: 430 KDa / Theoretical: 430 KDa |
-Macromolecule #1: RNA polymerase
| Macromolecule | Name: RNA polymerase / type: protein_or_peptide / ID: 1 / Name.synonym: RNAP / Details: RNAP with YkzG-GFP fusion / Number of copies: 1 / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Experimental: 430 KDa / Theoretical: 430 KDa |
| Recombinant expression | Organism:  |
-Experimental details
-Structure determination
| Method | negative staining |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1 mg/mL |
|---|---|
| Buffer | pH: 7.8 Details: 20 mM Tris-HCl , 150 mM NaCl, 10 mM MgCl2, 5% (v/v) glycerol and 1 mM DTT |
| Staining | Type: NEGATIVE Details: The specimens were then stained with a 1 % (w/v) uranyl formate aqueous solution. |
| Vitrification | Cryogen name: NONE / Instrument: OTHER |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI F30 |
|---|---|
| Date | Dec 1, 2009 |
| Image recording | Category: CCD / Film or detector model: GATAN ULTRASCAN 4000 (4k x 4k) / Number real images: 260 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Sample stage | Specimen holder model: JEOL |
| Experimental equipment |  Model: Tecnai F30 / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Applied symmetry - Point group: C1 (asymmetric) / Resolution.type: BY AUTHOR / Resolution: 24.0 Å / Resolution method: OTHER / Software - Name: EMAN2.8 / Number images used: 10104 |
|---|
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)