[English] 日本語
 Yorodumi
Yorodumi- EMDB-2630: Structural Studies on the Authentic Mumps Virus Nucleocapsid Show... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-2630 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structural Studies on the Authentic Mumps Virus Nucleocapsid Showing Uncoiling by the Phosphoprotein | |||||||||
 Map data Map data | Mumps Nucleocapsid | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | mumps / virus / nucleocapsid / phosphoprotein / replication | |||||||||
| Function / homology |  Function and homology information Function and homology informationnegative stranded viral RNA transcription / negative stranded viral RNA replication / helical viral capsid / viral nucleocapsid / host cell cytoplasm / ribonucleoprotein complex / structural molecule activity / RNA binding Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | helical reconstruction / cryo EM / Resolution: 18.1 Å | |||||||||
 Authors Authors | Cox R / Pickar A / Qiu S / Tsao J / Rodenburg CM / Dokland T / Elson A / He B / Luo M | |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2014 Journal: Proc Natl Acad Sci U S A / Year: 2014Title: Structural studies on the authentic mumps virus nucleocapsid showing uncoiling by the phosphoprotein. Authors: Robert Cox / Adrian Pickar / Shihong Qiu / Jun Tsao / Cynthia Rodenburg / Terje Dokland / Andrew Elson / Biao He / Ming Luo /  Abstract: Mumps virus (MuV) is a highly contagious pathogen, and despite extensive vaccination campaigns, outbreaks continue to occur worldwide. The virus has a negative-sense, single-stranded RNA genome that ...Mumps virus (MuV) is a highly contagious pathogen, and despite extensive vaccination campaigns, outbreaks continue to occur worldwide. The virus has a negative-sense, single-stranded RNA genome that is encapsidated by the nucleocapsid protein (N) to form the nucleocapsid (NC). NC serves as the template for both transcription and replication. In this paper we solved an 18-Å-resolution structure of the authentic MuV NC using cryo-electron microscopy. We also observed the effects of phosphoprotein (P) binding on the MuV NC structure. The N-terminal domain of P (PNTD) has been shown to bind NC and appeared to induce uncoiling of the helical NC. Additionally, we solved a 25-Å-resolution structure of the authentic MuV NC bound with the C-terminal domain of P (PCTD). The location of the encapsidated viral genomic RNA was defined by modeling crystal structures of homologous negative strand RNA virus Ns in NC. Both the N-terminal and C-terminal domains of MuV P bind NC to participate in access to the genomic RNA by the viral RNA-dependent-RNA polymerase. These results provide critical insights on the structure-function of the MuV NC and the structural alterations that occur through its interactions with P. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_2630.map.gz emd_2630.map.gz | 8.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-2630-v30.xml emd-2630-v30.xml emd-2630.xml emd-2630.xml | 8 KB 8 KB | Display Display |  EMDB header EMDB header |
| Images |  EMD-2630.png EMD-2630.png | 179.6 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-2630 http://ftp.pdbj.org/pub/emdb/structures/EMD-2630 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-2630 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-2630 | HTTPS FTP |
-Validation report
| Summary document |  emd_2630_validation.pdf.gz emd_2630_validation.pdf.gz | 235.7 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_2630_full_validation.pdf.gz emd_2630_full_validation.pdf.gz | 234.8 KB | Display | |
| Data in XML |  emd_2630_validation.xml.gz emd_2630_validation.xml.gz | 5.9 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-2630 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-2630 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-2630 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-2630 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_2630.map.gz / Format: CCP4 / Size: 29.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_2630.map.gz / Format: CCP4 / Size: 29.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Mumps Nucleocapsid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.29 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
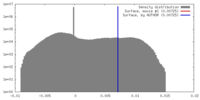
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Authentic mumps virus nucleocapsid
| Entire | Name: Authentic mumps virus nucleocapsid |
|---|---|
| Components |
|
-Supramolecule #1000: Authentic mumps virus nucleocapsid
| Supramolecule | Name: Authentic mumps virus nucleocapsid / type: sample / ID: 1000 / Oligomeric state: helical / Number unique components: 1 |
|---|
-Supramolecule #1: Mumps virus
| Supramolecule | Name: Mumps virus / type: virus / ID: 1 / NCBI-ID: 11161 / Sci species name: Mumps virus / Sci species strain: Iowa / Virus type: VIRION / Virus isolate: STRAIN / Virus enveloped: Yes / Virus empty: No |
|---|---|
| Host (natural) | Organism:  Homo sapiens (human) / synonym: VERTEBRATES Homo sapiens (human) / synonym: VERTEBRATES |
| Host system | Organism:  Chlorocebus aethiops (grivet) / Recombinant cell: vero Chlorocebus aethiops (grivet) / Recombinant cell: vero |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | helical array |
- Sample preparation
Sample preparation
| Vitrification | Cryogen name: ETHANE / Instrument: FEI VITROBOT MARK I |
|---|
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI F20 |
|---|---|
| Date | Apr 11, 2013 |
| Image recording | Category: CCD / Film or detector model: GATAN ULTRASCAN 4000 (4k x 4k) |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: OTHER |
| Sample stage | Specimen holder model: GATAN LIQUID NITROGEN |
| Experimental equipment |  Model: Tecnai F20 / Image courtesy: FEI Company |
- Image processing
Image processing
| Details | IHRSR using SPARX/EMAN2 package |
|---|---|
| Final reconstruction | Applied symmetry - Helical parameters - Δz: 5.32 Å Applied symmetry - Helical parameters - Δ&Phi: 28.3 ° Applied symmetry - Helical parameters - Axial symmetry: C1 (asymmetric) Resolution.type: BY AUTHOR / Resolution: 18.1 Å / Resolution method: OTHER / Software - Name: SPARX |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)