+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-2574 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | SA11 Rotavirus trypsinized Triple Layered Particle | |||||||||
 Map data Map data | 3DR of tripsinized SA11 strain rotacirus TLP | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | virus assembly / rotavirus / Proteolysis | |||||||||
| Biological species |   Simian rotavirus A/SA11 Simian rotavirus A/SA11 | |||||||||
| Method | single particle reconstruction / cryo EM / negative staining / Resolution: 14.5 Å | |||||||||
 Authors Authors | Rodriguez JM / Chichon FJ / Martin-Forero E / Gonzalez-Camacho F / Carrascosa JL / Caston JR / Luque D | |||||||||
 Citation Citation |  Journal: PLoS Pathog / Year: 2014 Journal: PLoS Pathog / Year: 2014Title: New insights into rotavirus entry machinery: stabilization of rotavirus spike conformation is independent of trypsin cleavage. Authors: Javier M Rodríguez / Francisco J Chichón / Esther Martín-Forero / Fernando González-Camacho / José L Carrascosa / José R Castón / Daniel Luque /  Abstract: The infectivity of rotavirus, the main causative agent of childhood diarrhea, is dependent on activation of the extracellular viral particles by trypsin-like proteases in the host intestinal lumen. ...The infectivity of rotavirus, the main causative agent of childhood diarrhea, is dependent on activation of the extracellular viral particles by trypsin-like proteases in the host intestinal lumen. This step entails proteolytic cleavage of the VP4 spike protein into its mature products, VP8* and VP5*. Previous cryo-electron microscopy (cryo-EM) analysis of trypsin-activated particles showed well-resolved spikes, although no density was identified for the spikes in uncleaved particles; these data suggested that trypsin activation triggers important conformational changes that give rise to the rigid, entry-competent spike. The nature of these structural changes is not well understood, due to lack of data relative to the uncleaved spike structure. Here we used cryo-EM and cryo-electron tomography (cryo-ET) to characterize the structure of the uncleaved virion in two model rotavirus strains. Cryo-EM three-dimensional reconstruction of uncleaved virions showed spikes with a structure compatible with the atomic model of the cleaved spike, and indistinguishable from that of digested particles. Cryo-ET and subvolume average, combined with classification methods, resolved the presence of non-icosahedral structures, providing a model for the complete structure of the uncleaved spike. Despite the similar rigid structure observed for uncleaved and cleaved particles, trypsin activation is necessary for successful infection. These observations suggest that the spike precursor protein must be proteolytically processed, not to achieve a rigid conformation, but to allow the conformational changes that drive virus entry. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_2574.map.gz emd_2574.map.gz | 165.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-2574-v30.xml emd-2574-v30.xml emd-2574.xml emd-2574.xml | 8.6 KB 8.6 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_2574.jpg emd_2574.jpg | 161.5 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-2574 http://ftp.pdbj.org/pub/emdb/structures/EMD-2574 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-2574 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-2574 | HTTPS FTP |
-Validation report
| Summary document |  emd_2574_validation.pdf.gz emd_2574_validation.pdf.gz | 306.1 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_2574_full_validation.pdf.gz emd_2574_full_validation.pdf.gz | 305.3 KB | Display | |
| Data in XML |  emd_2574_validation.xml.gz emd_2574_validation.xml.gz | 7.1 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-2574 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-2574 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-2574 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-2574 | HTTPS FTP |
-Related structure data
| Related structure data |  2573C  2575C  2576C  2577C  2578C  2579C  2580C C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_2574.map.gz / Format: CCP4 / Size: 175.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_2574.map.gz / Format: CCP4 / Size: 175.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | 3DR of tripsinized SA11 strain rotacirus TLP | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.8 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
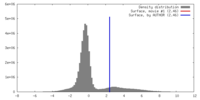
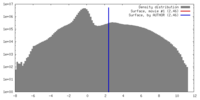
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Trypsinized Triple Layered Particle from SA11 strain rotavirus
| Entire | Name: Trypsinized Triple Layered Particle from SA11 strain rotavirus |
|---|---|
| Components |
|
-Supramolecule #1000: Trypsinized Triple Layered Particle from SA11 strain rotavirus
| Supramolecule | Name: Trypsinized Triple Layered Particle from SA11 strain rotavirus type: sample / ID: 1000 / Oligomeric state: icosahedral / Number unique components: 1 |
|---|
-Supramolecule #1: Simian rotavirus A/SA11
| Supramolecule | Name: Simian rotavirus A/SA11 / type: virus / ID: 1 / NCBI-ID: 10923 / Sci species name: Simian rotavirus A/SA11 / Sci species strain: SA11 / Virus type: VIRION / Virus isolate: STRAIN / Virus enveloped: No / Virus empty: No |
|---|---|
| Host (natural) | Organism:  Simiiformes (mammal) / synonym: VERTEBRATES Simiiformes (mammal) / synonym: VERTEBRATES |
-Experimental details
-Structure determination
| Method | negative staining, cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 Details: Tris 25 mM, Na2HPO3 0.7 mM, NaCl 136.9 mM, KCl 5.1 mM, Glucose 5.6 mM, MgCl2 1 mM, CaCl2 1 mM |
|---|---|
| Staining | Type: NEGATIVE Details: Samples were applied to grids, blotted and plunged into liquid ethane |
| Grid | Details: R 2/2 Quantifoil grids |
| Vitrification | Cryogen name: ETHANE / Instrument: OTHER |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI F20 |
|---|---|
| Date | Feb 1, 2011 |
| Image recording | Digitization - Scanner: ZEISS SCAI / Digitization - Sampling interval: 7 µm / Number real images: 108 / Average electron dose: 10 e/Å2 / Bits/pixel: 8 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 50000 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.26 mm / Nominal defocus max: 3.5 µm / Nominal defocus min: 0.75 µm / Nominal magnification: 50000 |
| Sample stage | Specimen holder: Eucentric / Specimen holder model: GATAN LIQUID NITROGEN |
| Experimental equipment |  Model: Tecnai F20 / Image courtesy: FEI Company |
- Image processing
Image processing
| CTF correction | Details: Phase flipping & amplitude decay |
|---|---|
| Final reconstruction | Applied symmetry - Point group: I (icosahedral) / Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 14.5 Å / Resolution method: OTHER / Software - Name: Xmipp / Number images used: 2682 |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)